Midterm 2 review
Midterm 2 Review Session
Midterm 2:
- Date: Thursday, November 23
- Duration: 80 minutes (5:00 - 6:20 PM)
- Location: same place as lecture (EME 0050)
- Mode: In-person - NO online option is available
- Coverage: Lectures 6-14 (
Lecture 15 has been removed from the testable material) and Labs 4-8 inclusive. (i.e. classification methods/methods, resampling techniques, and trees)
Format
- On paper - handwritten.
- a mix of multiple choice, and short/long answer
- closed-book, closed-technology
- i.e. computers and calculators are not permitted
- 1 doubled-sided 8.5x11 handwritten formula sheet is allowed
Lecture 6: Logistic Regression
Logistic Regression [slides]
Classification: Classification is a process of categorizing items or data into different classes or groups based on their characteristics or attributes.
Example: We may wish to investigate how variables, such as GRE (Graduate Record Exam scores), GPA (grade point average) and prestige of the undergraduate institution, effect admission into graduate school. The response variable, admit/don’t admit, is a binary variable. Hence this would be a binary classification problem.
Multiple Linear Regression
Recall the linear regression model: \[\begin{equation} Y = \beta_0 + \beta_1 X_1 + \cdots + \beta_p X_p + \epsilon \end{equation}\]
- \(Y\) is the random, quantitative response variable
- \(X_j\) is the \(j^{th}\) random predictor variable
- \(\beta_0\) is the intercept
- \(\beta_j\) from \(j = 1, \dots p\) are the regression coefficients
- \(\epsilon\) is the error term
Generalized Linear Model
Rather than modeling continuous \(Y \in (-\infty, \infty)\) using: \[\begin{equation} Y = \underbrace{\beta_0 + \beta_1 X_1 + \cdots + \beta_p X_p}_{\eta} \end{equation}\]
A GLM consists of three components:
- Systematic component
- Random component
- Link function
Link Function:
In statistics, a link function is a mathematical function that connects the linear predictor of a statistical model to the expected value of the response variable.
\[\begin{align*} g(\eta) &= \mu \end{align*}\]where \(\mu = {E}[Y_i]\). In logistic regression, \(Y_i \sim \text{Bernoulli}(\pi_i)\) and \(E[Y_i] = \pi_i\), i.e. the probability \(Y = 1\). So we have:
\[ \Pr(Y = 1 \mid X = x) = g(\eta) \]
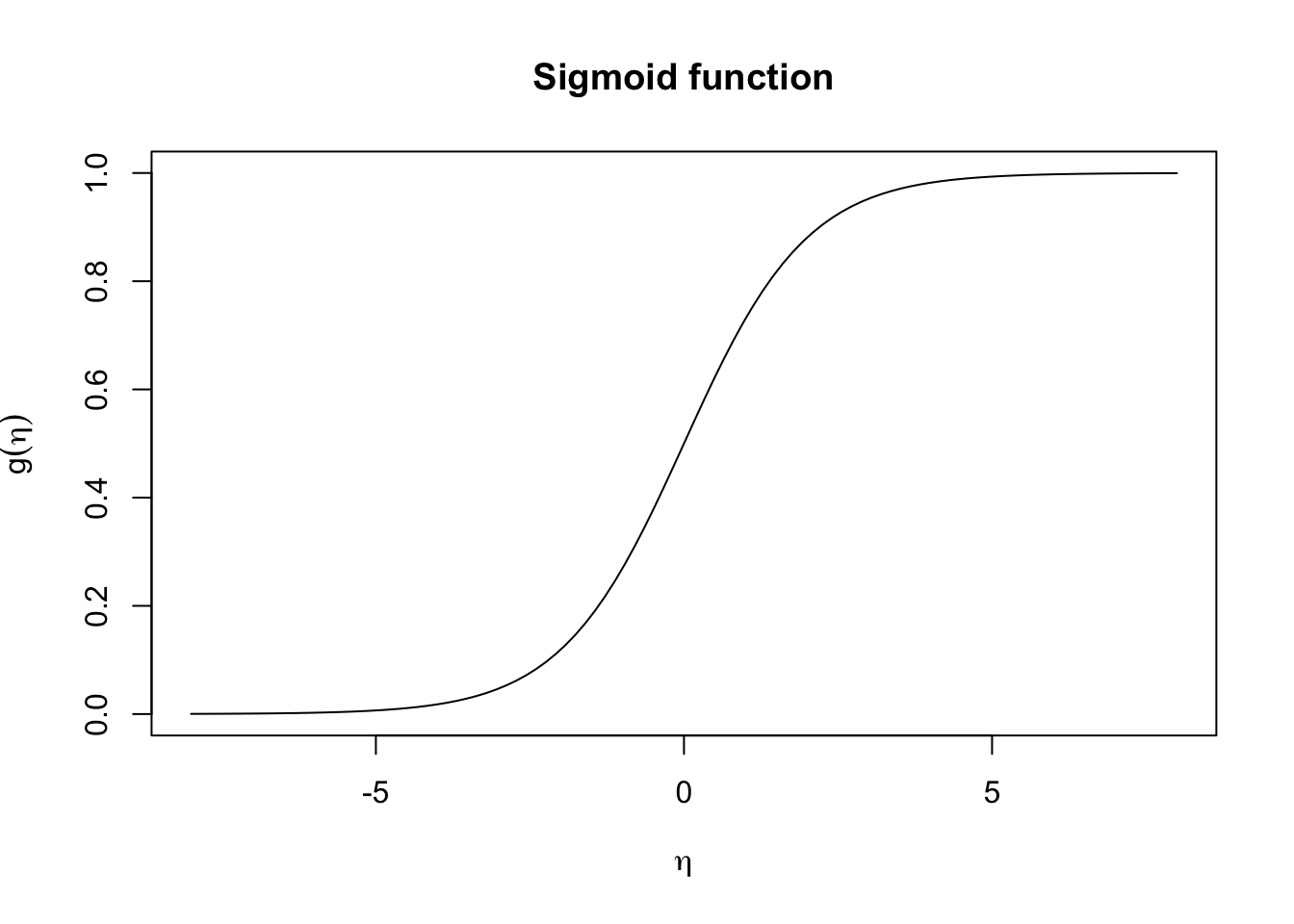
In logistic regression we use the logistic link function \(g(x) = \frac{1}{1 + e^{-x}}\) (i.e. sigmoid function) to map the linear combination of predictors, \(\eta\) to probabilities, where
\[\begin{align*} g(\eta) &= \Pr(Y = 1 \mid X = x) \\ &= \dfrac{1}{1 + e^{-\eta}}\\ &= \dfrac{1}{1+ e^{-(\beta_0 + \beta_1 X_1 + \cdots \beta_p X_p)}} \end{align*}\]In other words, this will transforms the linear predictor \(\eta = \beta_0 + \beta_1 X_1 + \cdots \beta_p X_p\) into a value between 0 and 1.
Alternatively you can think of this in terms of \(g^{-1}()\), i.e., the inverse link function, which is the logit function, \(\text{logit}(p) = \ln \left( \frac{p}{1-p} \right)\) in the case of logistic regression.
\[ \begin{align*} g(\eta) &= \mu \\ \implies \eta &= g^{-1}(\mu) \\ &= \ln \left( \frac{\mu}{1-\mu} \right) \end{align*} \]
In other words, the linear predictor \(\eta = \beta_0 + \beta_1 x_1 + \cdots \beta_p x_p\) provides the log-odds of \(Y = 1\) given some particular set of \((x_1, x_2, \dots, x_p)\) values.
Logistic Regression:
\[ \text{log odds} = \beta_0 + \beta_1 X_1 + \dots + \beta_p X_p \]
Assumptions
\[ \begin{align} Y_i &\sim \text{Bernoulli}(\pi_i) = \mu \\ g( \eta) & = \dfrac{1}{1+e^{-\sum_{j=1}^p\beta_j X_j}} \end{align} \]
where \(g(\cdot)\) is the logistic function. Note: parameters are fit by maximum likelihood estimation (MLE) rather than ordinary least squares (OLS).
Interpretation of Coefficients
Since probability \(\pi_i\) is non-linear as \(X\) varies1, we cannot interpret the coefficients as we did in regular regression. The best we can do is talk about the direction:
Holding all other variables constant, if \(\beta_j\) is positive, then an increase in \(X_j\) is associated with an increase in \(\pi_i\)
Holding all other variables constant, if \(\beta_j\) is negative, then an increase in \(X_j\) is associated with a decrease in \(\pi_i\).
R code
You can fit a logistic regression using the glm() function.
glm(formula, family = gaussian, data, ...)| Arguments | Description |
|---|---|
formula |
a symbolic description of the model to be fitted, eg Y~ X1 + X2 |
family |
a description of the error distribution and link function to be used in the model ?family |
data |
data frame (usually your training set) |
To perform logistic regression with a binary outcome we need to set family = “binomial”. Other options (not be covered in this course) include:
family = "gaussian"same aslm()family = "poisson"for predicting countsfamily = multinomialfor \(>2\) classesfamily = binomial('probit')probit regression (a common alternative to logistic regression)
A note about factor levels
Note that logistics regression takes the second level of a factor to be the \(Y = 1\).
Example
Using a single predictor
library(gclus)
data(body)
attach(body)
body$Gender <- factor(Gender)
simlog <- glm(Gender ~ Height , family="binomial", data = body)
# store the beta hats for easy referencing
betas <- coef(simlog)
# plot the data (renaming the y-axis)
plot(Gender~Height, ylab="Probility of being Male")
# plot the fitted curve p(x) = e^(b0+b2x)/(1 + e^(b0+b2x))
curve(
(exp(betas[1] + betas[2]*x))/(1+exp(betas[1] + betas[2]*x)),
add=TRUE, # superimposes onto scatterplot (otherwise new plot)
lwd=2, # line width (make the line a bit thicker)
col=2) # 2 = red 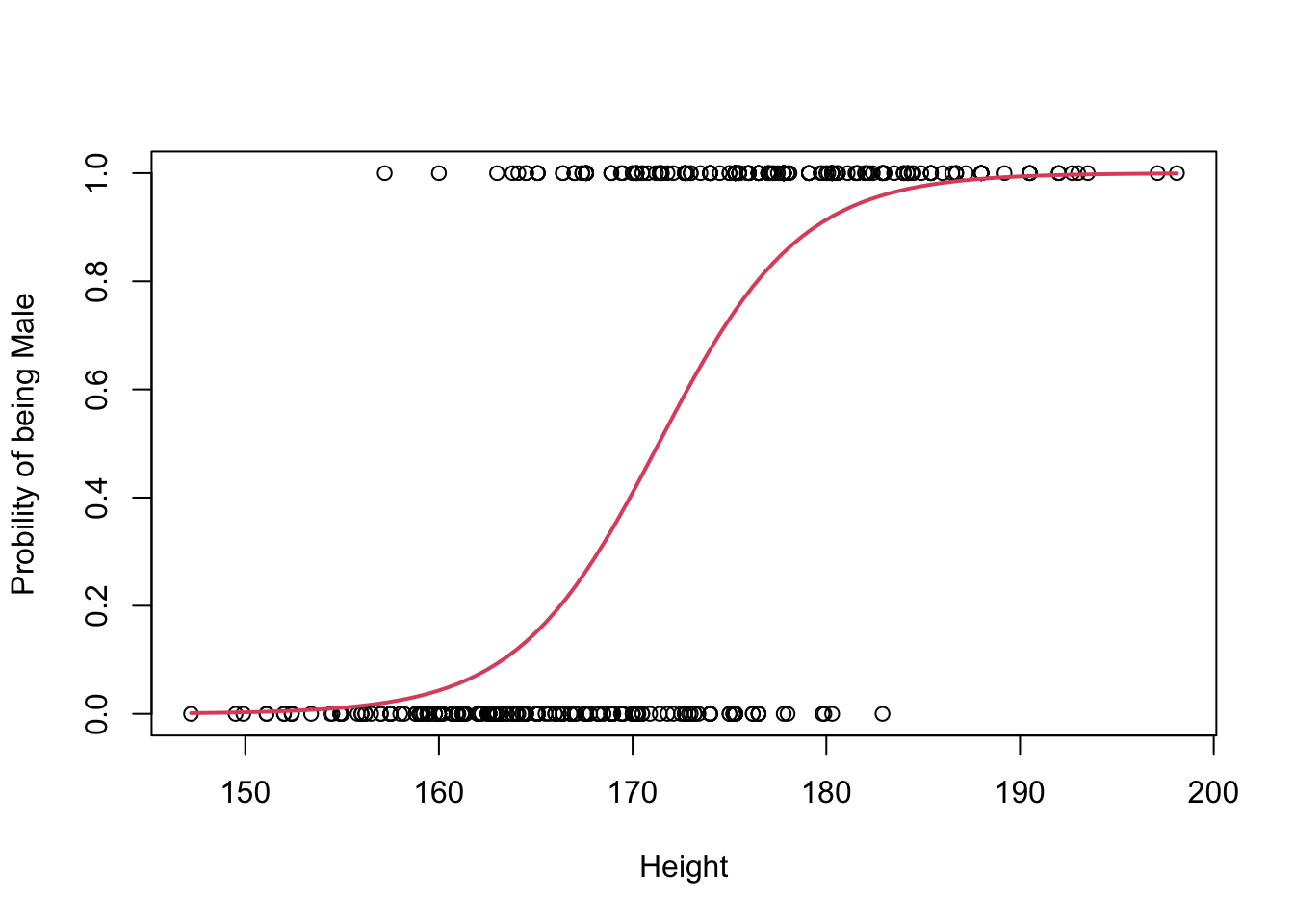
On paper we can calculate the \(\Pr(Y = 1 \mid X = x)\) using the formula
\[\begin{align*} \Pr(Y = y \mid X = 175) &= \dfrac{1}{1+ e^{-(\beta_0 + \beta_1 X_1 )}}\\ &= \dfrac{1}{1+ e^{-(-46.7632791 + 0.2729192 (175))}}\\ &= 0.7305827 \end{align*}\]In R we could obtain these predictions using the predict() function:
newpt = data.frame(Height = 175)
predict(simlog, newdata = newpt, type="response") 1
0.7305827 We can also find the Height value that gives the linear boundary between the two prediction regions.
\[ \begin{align} \Pr(Y = y \mid \texttt{Height} = x) &= \dfrac{1}{1+ e^{-(\beta_0 + \beta_1 X_1 )}}\\ 0.5 &= \dfrac{1}{1+ e^{-(\beta_0 + \beta_1 X_1 )}} \\ 1 &= 0.5 + 0.5 \times e^{-(\beta_0 + \beta_1 X_1 )}\\ \frac{0.5}{0.5} &= e^{-(\beta_0 + \beta_1 X_1 )}\\ \log \left(1\right) &= {\beta_0 + \beta_1 X} \\ \implies X &= -\frac{\beta_0}{\beta_1} \end{align} \] So for Height \(> -\dfrac{ -46.7632791}{0.2729192} = 171.34\) we would predict Male.
Using multiple predictors
# Fit the logistic regression model
mlr <- glm(factor(Gender) ~ Height + WaistG + BicepG, family="binomial")
summary(mlr)
Call:
glm(formula = factor(Gender) ~ Height + WaistG + BicepG, family = "binomial")
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -55.32670 5.51960 -10.024 < 2e-16 ***
Height 0.21534 0.02864 7.520 5.50e-14 ***
WaistG 0.05167 0.02891 1.787 0.0739 .
BicepG 0.46541 0.08265 5.631 1.79e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 702.52 on 506 degrees of freedom
Residual deviance: 232.90 on 503 degrees of freedom
AIC: 240.9
Number of Fisher Scoring iterations: 6In this case, larger predictor values would result in higher probabilities that the observation is male.
Lecture 7: Assessing Classification Models
We often look at confusion matrices that have the form:
Confusion Matrix
| Predicted Yes | Predicted No | |
|---|---|---|
| Actual Yes | TP = True Positive | FN = False Negative |
| Actual No | FP = False Positive | TN= True Negative |
Primary Measures
From the confusion matrix we can extract things like:
Misclassifcation (aka error) rate: \(\frac{FP + FN}{\text{Total number of predictions}}\)
Accuracy: \(\frac{TP + TN}{\text{Total number of prediction}}\)
Accuracy can sometimes be a misleading measure (especially with umbalanced2 data). Other metrics we can consider are:
Precision: \(\frac{TP}{TP + FP}\) = proportion of predicted Yes’ that are actually Yes.
- e.g. good for spam detection
Recall (aka sensitivity): \(\frac{TP}{TP + FN}\) the proportion of actual Yes’ that were predicted Yes.
- e.g good for disease diagnostic
Specificity: \(\frac{TN}{TN + FP}\) the proportion of actual No’s that were predicted No.
- e.g. good for quality control
F1
Another useful/common metric is the F1 score. It is the harmonic mean between Precision and Recall.
\[ \text{F1} = \dfrac{2 \times \text{Precision} \times \text{Recall}} {\text{Precision} + \text{Recall}} \]
This is a value between 0 and 1 (0 being the worst score, 1 being the best score). In practical scenarios, the F1 score typically falls between 0 and 1. Values closer to 1 indicate better balance between precision and recall, while values closer to 0 suggest an imbalance or trade-off between precision and recall.
Note: while the F1 score is simplest to explain in the binary classification problem, it can be used in the multiclass scenario as well (details not provided).
The harmonic mean is particularly useful in situations where extreme values (e.g., one of precision or recall being very low) should heavily impact the overall score.
# Example data
precision <- 0.9
recall <- 0.1
hmean <- function(p,f) { 2*p*f/(p+f)}
# Calculate the harmonic mean (F1 score)
hmean(precision, recall)[1] 0.18mean(c(precision, recall))[1] 0.5F1 score is at its minimum when either precision or recall is zero.
- This occurs when all predictions are incorrect for one of the classes (e.g., no true positives for recall or no true positives for precision). In such cases, the F1 score approaches 0.
The F1 score is at its maximum value of 1 when both precision and recall are perfect.
- This means that the model has made no false positive or false negative predictions, achieving both high precision and high recall.
ROC
The Receiver Operating Characteristic (ROC) curve is a graphical representation of the performance of a binary classification model across various discrimination thresholds. It illustrates the trade-off between the true positive rate (sensitivity) and the false positive rate (1-specificity) as the decision threshold for classifying positive instances is varied.
The ROC curve is created by plotting the true positive rate (sensitivity) against the false positive rate (1-specificity) at various threshold values for the model’s predicted probabilities. Each point on the curve corresponds to a different decision threshold.
An ideal ROC curve will hug the top left corner, so the larger the AUC, the better the classifier.
Looking at out body example:
Code
library(ROCR)
truey = body$Gender
pp1 = predict(simlog, type = "response")
pp2 = predict(mlr, type = "response")
pred1 = prediction(pp1, truey)
pred2 = prediction(pp2, truey)
perf1 = performance(pred1, measure = "tpr", x.measure = "fpr")
perf2 = performance(pred2, measure = "tpr", x.measure = "fpr")
plot(perf1, col = "blue", lwd = 2, main = "ROC Curves", col.main = "black", lty = 1)
plot(perf2, col = "red", lwd = 2, add = TRUE)
legend("bottomright", lwd = 1, col = c("red", "blue"), legend = c("logisitic regression with multiple predictors (simlog)", "logistic regression with a single predictor (mlr)"))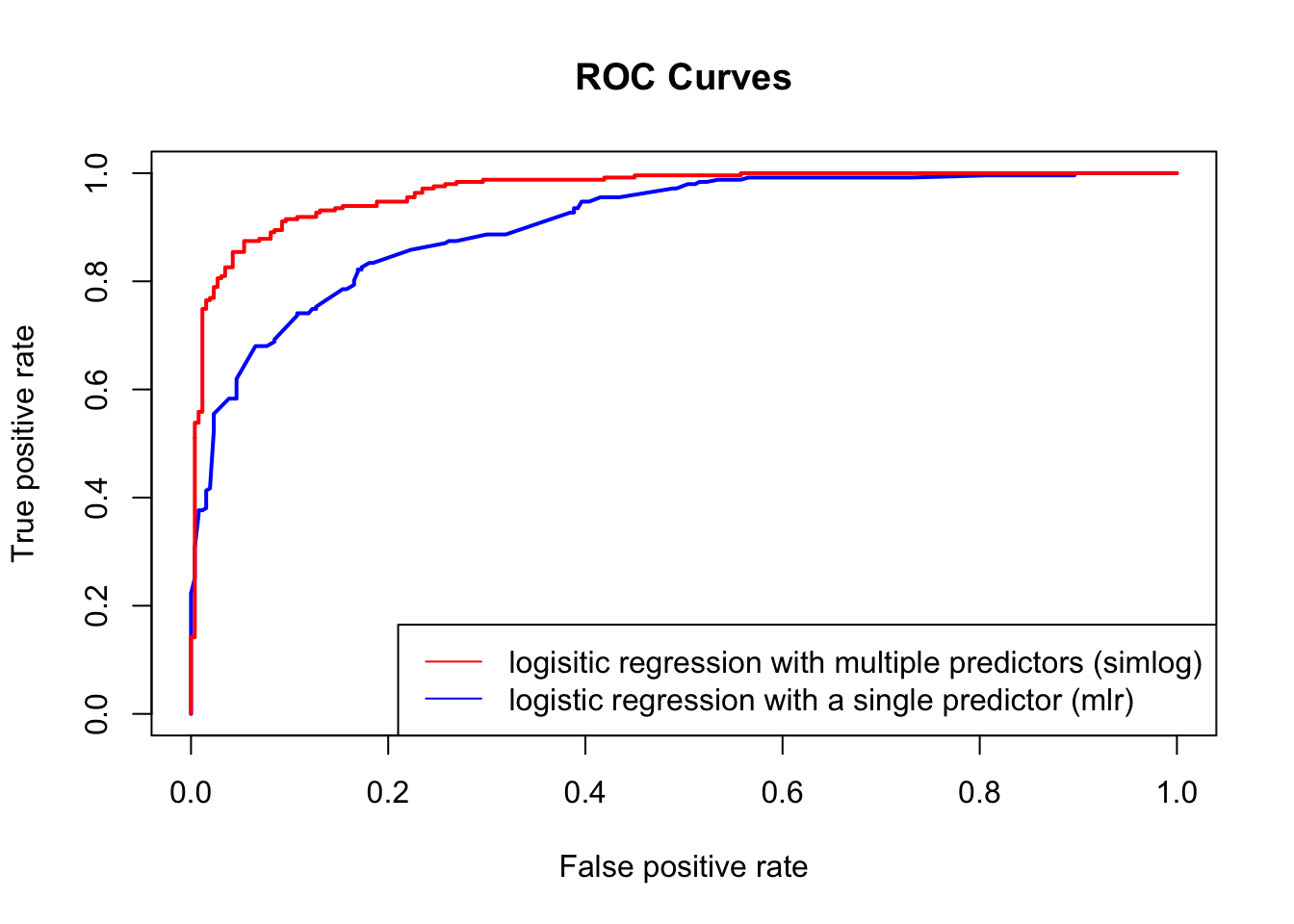
Code
library(pROC) # this is different than the one in lab but easier to use for areas
# Create a ROC curve object
roc_curve1 <- roc(truey, pp1)
roc_curve2 <- roc(truey, pp2)
# Get the AUC-ROC value
auc_value1 <- auc(roc_curve1)
auc_value2 <- auc(roc_curve2)Since the area under the red ROC curve (=0.9680785) is larger than the area under the blue ROC curve (=0.9072252), the the red model (i.e. mlr corresponding to logisitic regression with multiple predictors) would be deemed better than the blue model (i.e. simlog corresponding to logisitic regression with multiple predictors)
Logloss
The logloss function is a widely used for binary and multi-class classification problems. It measures the performance of a classification model by quantifying how well the predicted probabilities align with the true class labels. Log Loss is particularly useful in probabilistic models where predictions are expressed as probabilities. The definition is given by this (but I wouldn’t ask you to compute this on a midterm)
\[-\frac{1}{n} \sum_{i=1}^{n} \sum_{g=1}^G I(y_i = g) \log z_{ig}\]
- When the logloss = 0, the predicted probabilities for all instances perfectly match the true class probabilities.
- Log loss increases as the predicted probabilities deviate from the true class probabilities.
- Log loss has no specific upper bound.
There are various packages in R that will compute these metrics for us
Lecture 8: Distance Measures
Distance play a crucial role many machine learning models. In this lecture we explored a few differnt types of distance calculations
Euclidean Distance
The Euclidean distance between observations \(x_i = (x_{i1}, x_{i2}, \dots x_{ip})\) and \(x_j = (x_{j1}, x_{j2}, \dots x_{jp})\) is calculated as \[d^{\text{EUC}}_{ij} = \sqrt{\sum_{k=1}^p (x_{ik} - x_{jk})^2}\]
We often normalize (scale and center the data to have mean 0, variance 1) via \[z_{ik} = \frac{x_{ik}-\mu_k}{\sigma_k}\] then we can define standardized pairwise distances as \[d^{\text{STA}}_{ij} = \sqrt{\sum_{k=1}^p (z_{ik} - z_{jk})^2}\]
Manhattan Distance
Manhattan distance3 measures pairwise distance as though one could only travel along the axes \[d^{\text{MAN}}_{ij} = \sum_{k=1}^p |x_{ik} - x_{jk}|\]
Similarly, one might want to consider the standardized form \[d^{\text{MANs}}_{ij} = \sum_{k=1}^p |z_{ik} - z_{jk}|\]
Mahalanobis Distance
Mahalanobis distance takes into account the covariance structure of the data.
It is easiest defined in matrix form \[d^{\text{MAH}}_{ij} = \sqrt{(x_i - x_j)^T \boldsymbol{\Sigma}^{-1} (x_i - x_j)}\] where \(^T\) represents the transpose of a matrix and \(^{-1}\) denotes the inverse of matrix.
Matching Binary Distance
We can actually define another matching method that is equivalent to Manhattan/city-block,
\[d^{\text{MAT}}_{ij} = \text{\# of variables with opposite value}\]
Asymmetric Binary Distance
Asymmetric binary distance is a measure of dissimilarity between two binary vectors (vectors with elements of 0 or 1) where differences in the presence or absence of certain features are considered unequal. The example we saw in class disregards 0-0 matches.
\[d^{\text{ASY}}_{ij} = \frac{\text{\# of 0-1 or 1-0 pairs}}{\text{\# of 0-1, 1-0, or 1-1 pairs}}\]
Distance for Qualitative Variables
For categorical variables having \(>2\) levels, a common measure is essentially standardized matching once again. \[d^{\text{CAT}}_{ij} = 1 - \frac{u}{p}\]
where \(u\) is the number of variables that match between observations \(i\) and \(j\) and
\(p\) is the total number of variables
Gower’s Distance
Gower’s distance (or Gower’s dissimilarity) is calculated as: \[d^{\text{GOW}}_{ij} = \frac{\sum_{k=1}^p \delta_{ijk} d_{ijk}}{\sum_{k=1}^p \delta_{ijk}}\]
where
- \(\delta_{ijk}\) = 1 if both \(x_{ik}\) and \(x_{jk}\) are non-missing (0 otherwise),
- \(d_{ijk}\) depends on variable type
- Quantitative numeric \[d_{ijk} = \frac{|x_{ik} - x_{jk}|}{\text{range of variable k}}\]
- Qualitative (nominal, categorical) \[d_{ijk} = \begin{cases} 0 \text{ if obs i and j agree on variable k, } \\ 1 \text{ otherwise} \end{cases} \]
- Binary \(d_{ijk} = \begin{cases} 0 \text{ if obs i and j are a 1-1 match, } \\ 1 \text{ otherwise} \end{cases}\)
LDA and QDA
know the assumptions of each (and when to use which)
derivation of linear boundary
Lecture 10: Clustering
Unsupervised learning is utilized for three main tasks
- clustering
- dimensionality reduction
- association
Clustering is a type of unsupervised machine learning technique where the goal is to group similar data points into clusters or segments.
Hierarchical clustering
As its name suggests, hierarchical clustering is a type of clustering algorithm that builds a hierarchy of clusters.
There are two main types of hierarchical clustering:
Agglomerative (bottom-up approach) Hierarchical Clustering: starts with each data point as a separate cluster and then iteratively merges the closest clusters until only one cluster remains.
Divisive (top-down) Hierarchical Clustering: starts with all data points in a single cluster and then recursively splits the cluster into smaller clusters until each data point is in its own individual cluster.
We focus on agglomerative hierarchical clustering which takes the following steps:
Initialization: Start by treating each data point as a separate cluster. At the beginning, there are as many clusters as there are data points.
Compute Similarities/Distances: Calculate the pairwise distances or similarities between all clusters. The choice of distance metric depends on the nature of the data (e.g., Euclidean distance, Manhattan distance, etc.).
Merge Closest Clusters: Identify the two closest clusters based on the computed distances or similarities. The definition of “closeness” depends on the chosen distance metric.
Update Cluster Set:: Merge the two closest clusters into a single cluster, reducing the total number of clusters by one.
Repeat steps 2–4: Recalculate the distances or similarities between the updated clusters and repeat the process of identifying and merging the closest clusters. For this we must choose a linkage criteria (how to measure the distance between clusters). Common linkage methods include:
- Single linkage: calculates the minimum distance between two points in each cluster.
- Complete linkage: calculates the maximum distance between two points in each cluster.
- Average linkage: calculates the average pairwise distance between the observations inside group with those outside.
- Centroid Linkage: calculates the distance between the centroids (mean points) of two clusters.
In some cases, the choice of linkage method could make a big difference. (e.g. single linkage often leads to “chaining” which produces elongated clusters.
- Termination: Continue this process iteratively until only one cluster, containing all the data points, remains.
Don’t forget to normalize your data (we use the scale(x, center = TRUE, scale = TRUE) function for that)!
Dendrogram Construction:
Throughout the process, keep track of the merging order and heights at which merges occur. This information is used to construct a dendrogram, which visually represents the hierarchy of clusters.
data(faithful)
colors <- c("purple",
"dodgerblue",
"firebrick",
"forestgreen",
"gold")
fait <- hclust(dist(scale(faithful)), method="single")
plot(fait, main="", labels=FALSE)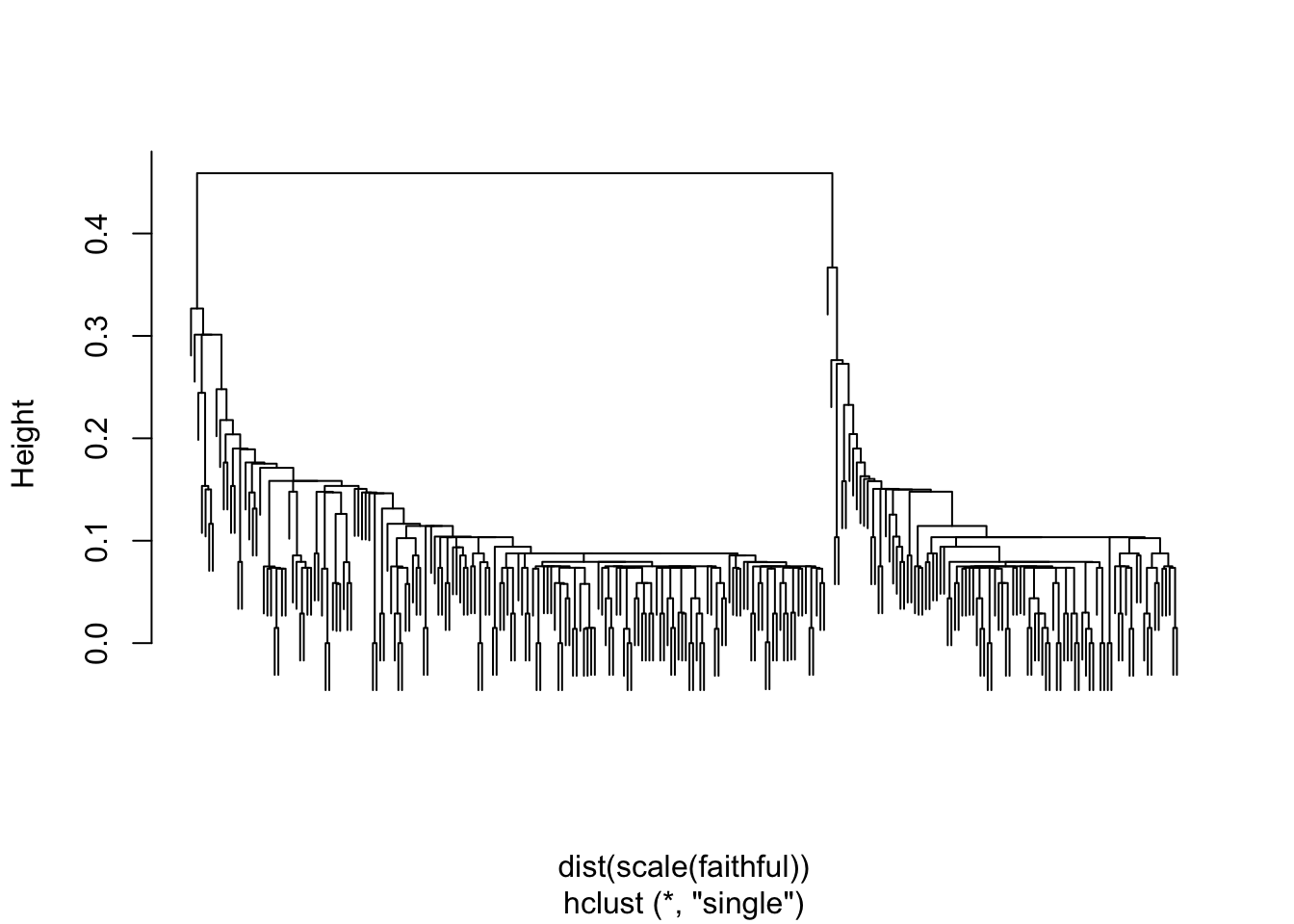
hc2 <- cutree(fait, k = 2)
plot(faithful, col = colors[hc2], pch=as.character(hc2))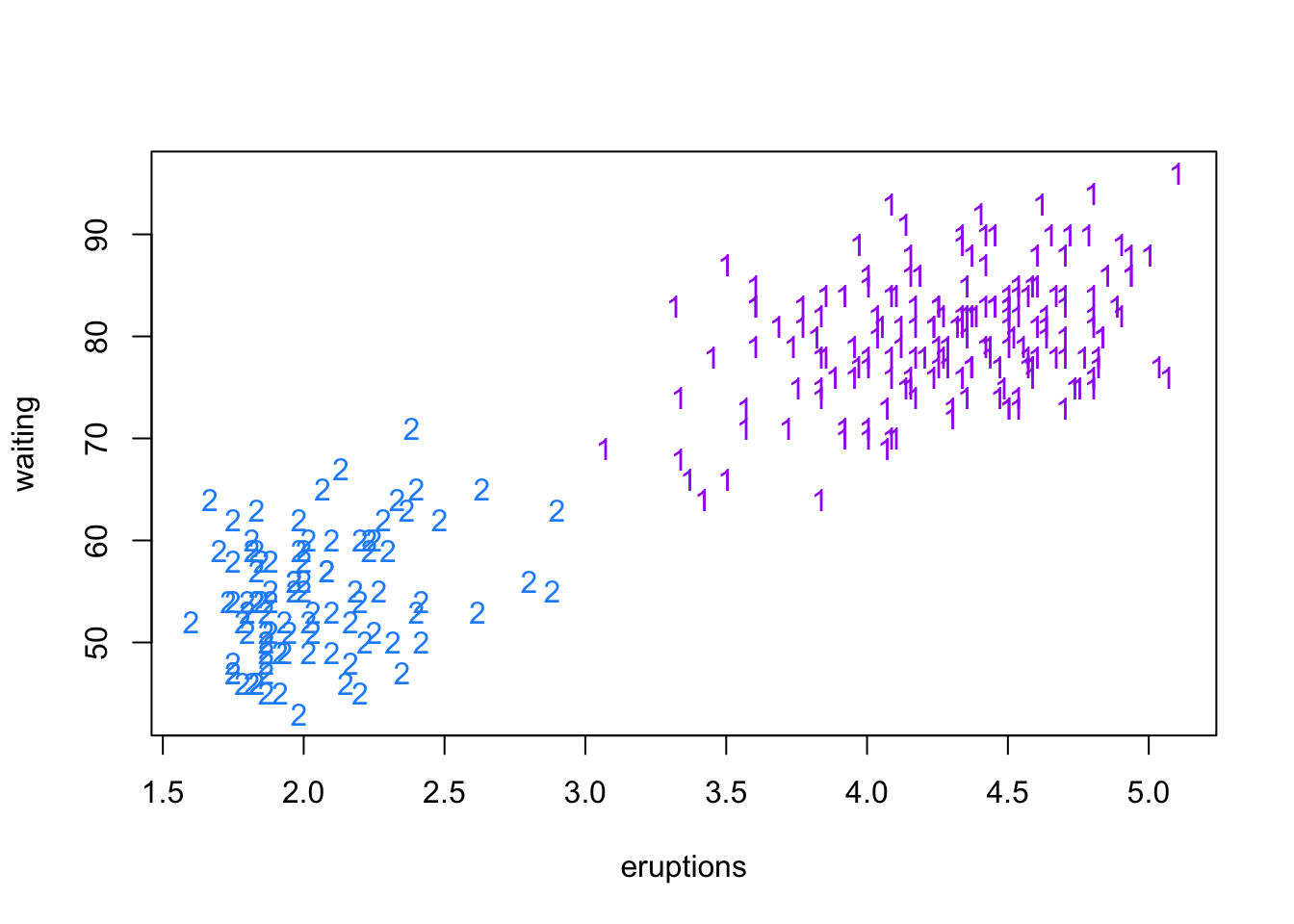
- Dendrograms can also be used to determine how many groups are present in the data.
- The number of clusters is determined by cutting the dendrogram at a certain height
- Generally, we can “cut” at the largest jump in distance, and that will determine the number of groups, as well as each observation’s group membership.
kmeans clustering
\(k\)-means clustering is a popular method for grouping n observations into \(k\) clusters; note that \(k\) needs to be provided by the user.
Algorithm
- Randomly select \(k\) (the number of groups) points in your data. These will serve as the first centroids4
- Assign all observations to their closest centroid (in terms of Euclidean distance). You now have \(k\) groups.
- Calculate the means of observations from each group; these are your new centroids.
- Repeat 2) and 3) until nothing changes anymore (each loop is called an iteration).
Objective: to minimize Within-Cluster-Sum of Squared Errors (WSS) , i.e. the sum of squared distances between the points and their respective cluster centroid.
Note that this alogrithm is stochastic (you may get a different answer based the random assignment in step 1.) To gain some stability we will often run the algorithm several times and pick the best one (in terms of WSS). For example, we can run kmeans 25 times using:
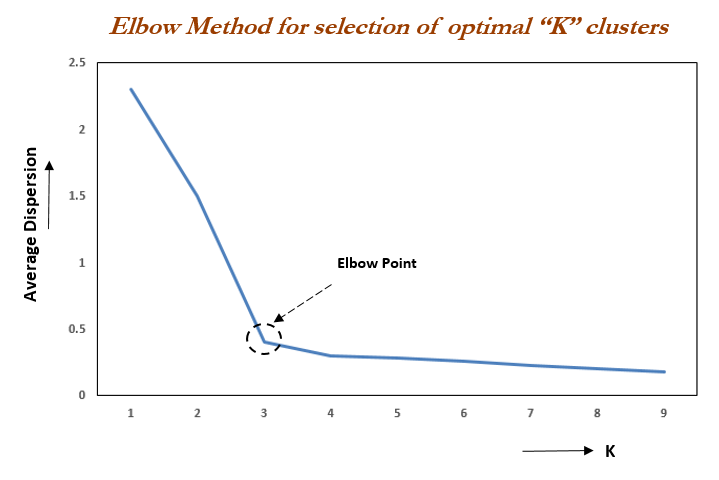
kmeans(x, centers=k, nstart = 25)Elbow method
To choose the value of \(k\) we can use the “elbow method”. It works as follows:
- Calculate the Within-Cluster-Sum of Squared Errors (WSS) for different values of \(k\),
- Choose the \(k\) for which WSS becomes first starts to diminish.
This appears int the WSS-versus-\(k\) plot (aka scree plot) in the form of an “elbow”.
Cross-validation
Lecture 11: Cross-validation
This method involves essentially preforming the holdout method iteratively with the training data.
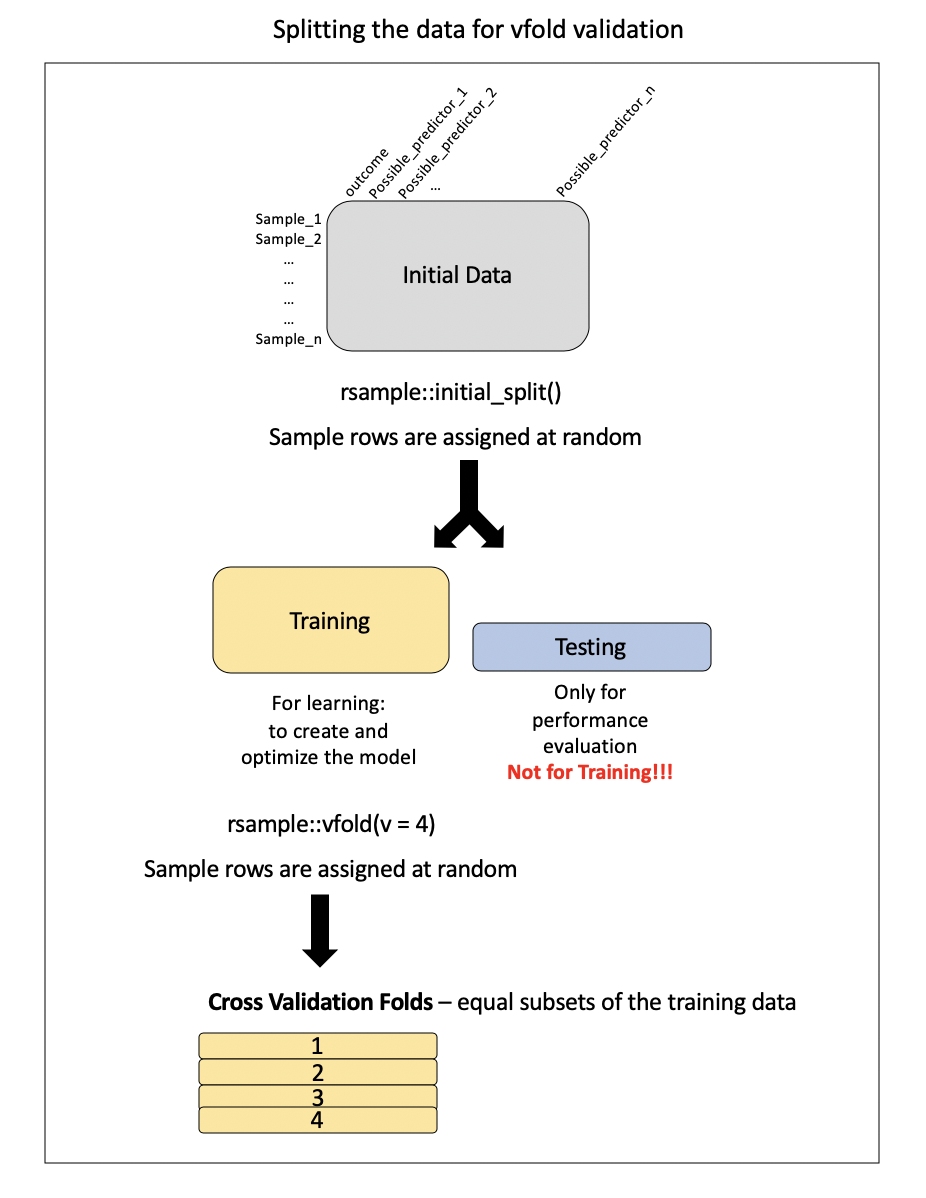
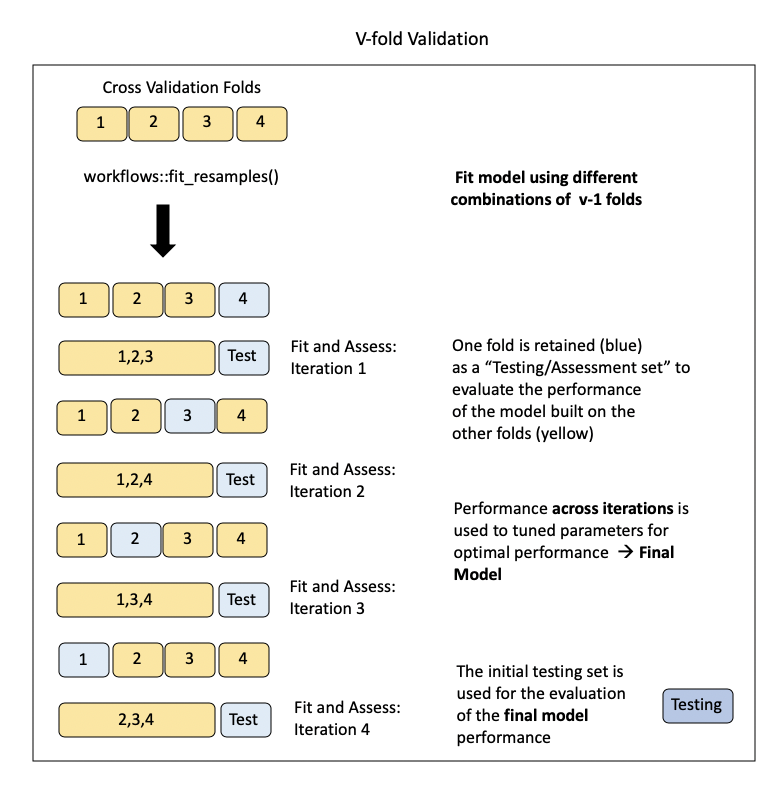
- LOOCV was a special case of k-fold cross-validation where \(k = n\)
- Usually 5- or 10-fold is used.
Pros of \(k\)-fold
- Less variance than LOOCV
- More computationally feasible than LOOCV (\(k\) vs. \(n\) model fits)
- Often gives more accurate estimates of test error than LOOCV.
Cons of \(k\)-fold
- Slightly more bias than towards overestimating MSE in comparison to LOOCV
- Non deterministic (randomly splitting the data into \(k\) folds)
Note: CV is often used to help select a model (also know as tuning a model). The actual reported model would be the one run with all the (training) data and reported evaluation metric would be that reported on from the test data.
Lecture 12: Bootstrap
Bootstrapping is a statistical procedure that resamples a single data set to create many simulated samples. This process allows for the calculation of standard errors, confidence intervals, and hypothesis testing
Both cross validation and bootstrapping are resampling methods.
- CV is often used to help select a model (also know as tuning a model)
- The bootstrap is often used to quantify the uncertainty associated with a given estimator or statistical learning method.
Steps
Consider some estimator \(\hat \alpha\). For \(j = 1, 2, \dots B\) 5
- take a random sample of size6 \(n\) from your original data set with replacement. This is the \(j\)th bootstrap sample, \(Z^{*j}\)
- calculate the estimated value of your parameter from your bootstrap sample, \(Z^{*j}\), call this value \(\hat \alpha^{*j}\).
Estimate the standard error and/or bias of the estimator, \(\hat \alpha\), using the equations given on the upcoming slides.
Sample with replacement
Like CV, this method will involve sampling from our data set, only now, we will be sampling with replacement.

Bootstrap Standard Error
Bootstrap estimates the standard error of estimator \(\hat \alpha\) using7
\[\begin{equation} \hat{\text{SE}}(\hat \alpha) = \sqrt{\frac{\sum_{i=1}^{B} (\hat \alpha^{*i} - \overline{\hat \alpha})^2}{B -1}} \end{equation}\]where \(\overline{\hat \alpha} = \sum_{j = 1}^B \hat \alpha^{*j}\) is the average estimated value over all \(B\) bootstrap samples.
Bootstrap Bias
The bootstrap estimates the Bias of estimator \(\hat \alpha\) using:
\[\begin{align} \hat{\text{Bias}}(\hat \alpha) = \overline {\hat \alpha} - \hat \alpha_f \end{align}\]where \(\overline{\hat \alpha} = \sum_{j = 1}^B \hat \alpha^{*j}\) and \(\hat \alpha_f\) is the estimate obtained on the original data set.
Bootstrap Confidence Intervals (CI)
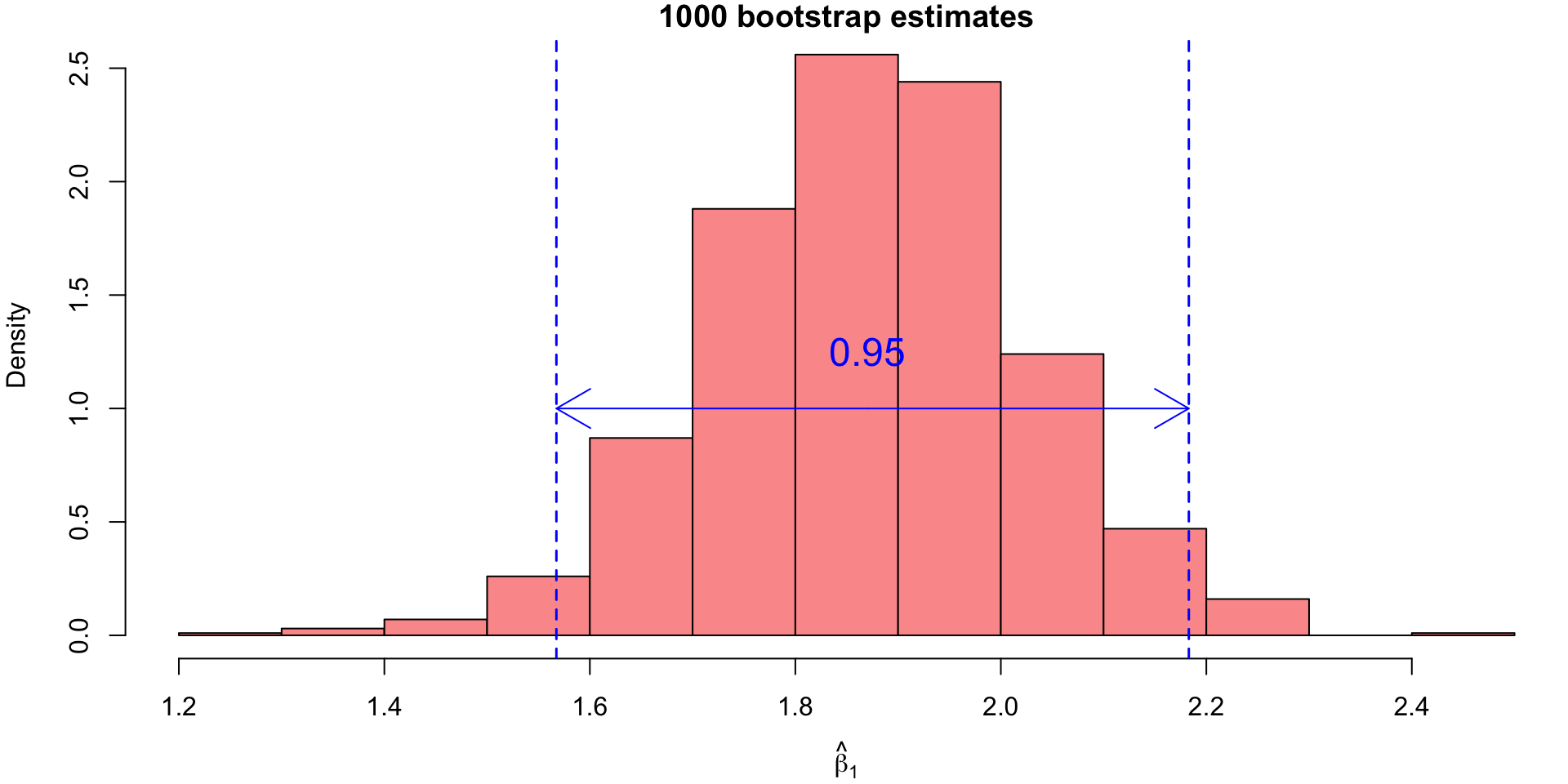
- To obtain confidence intervals from this empirical distribution, we would simply finding appropriate percentiles.
- For our example, the 95% bootstrap CI for \(\beta_1\) would correspond to the 25th value and the 975th value of the sorted bootstrap estimates.

Trees
CART < Bagging < Random Forests
Lecture 13: CART
Classification and regression trees: [slides]
Classification and Regression Trees (AKA CART or decision trees) involve partitioning (AKA stratifying or segmenting) the predictor/feature space by simple binary splitting rules.
Since the set of splitting rules used to segment the predictor space can be summarized in a tree, these types of approaches are known as decision tree methods.
We make a prediction for an obs. using the mean/mode response value for that “region” to which it belongs.
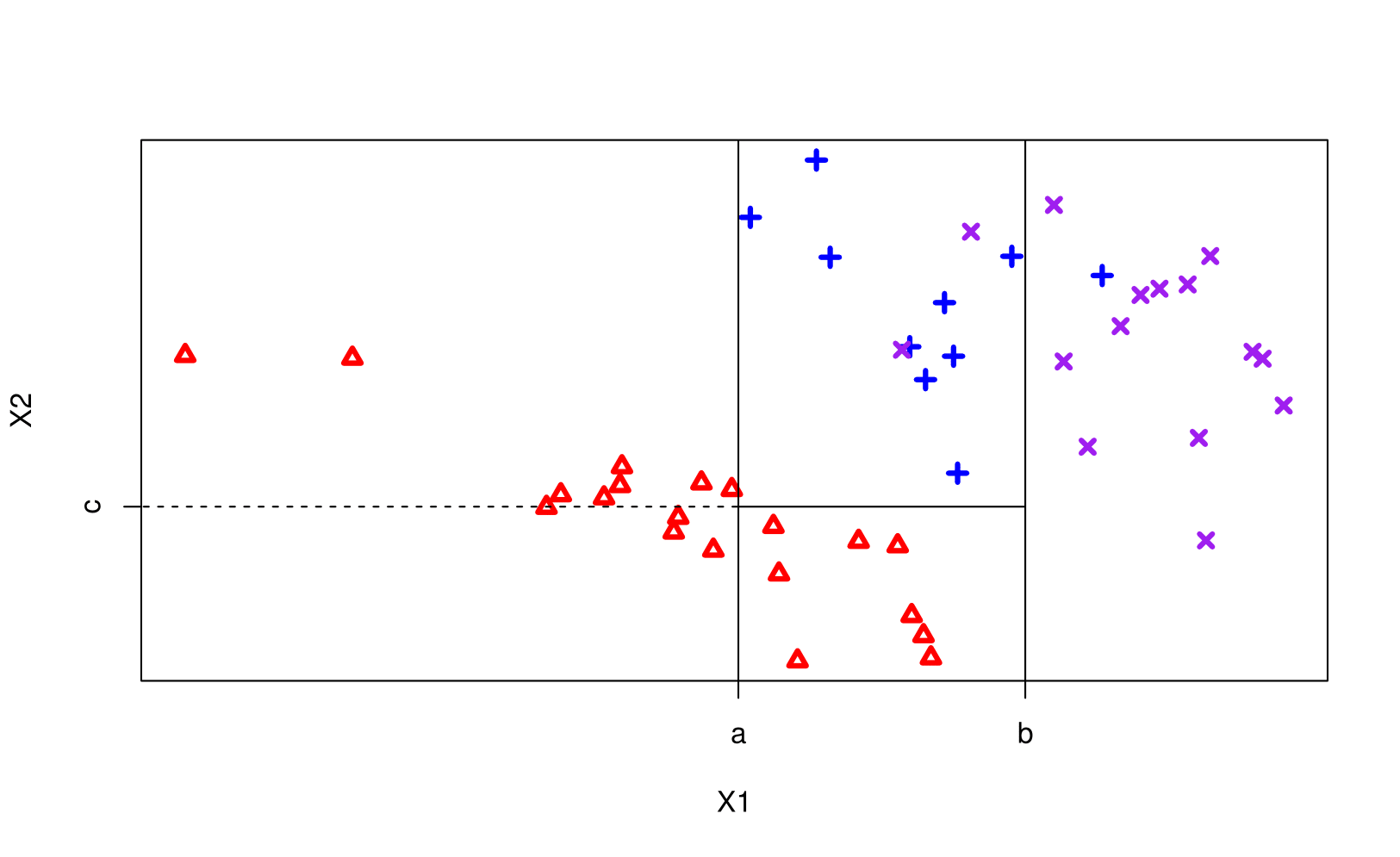
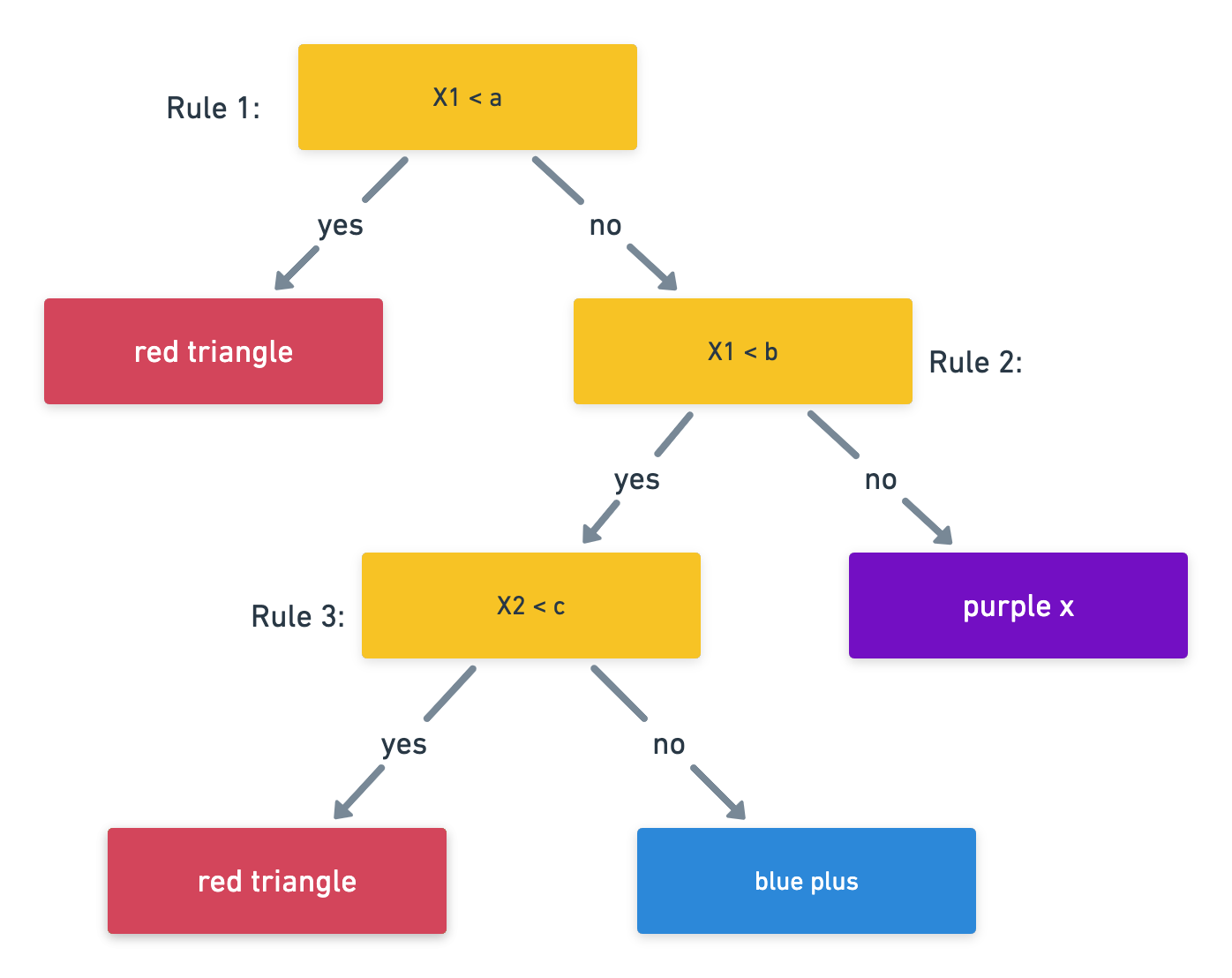
Notation:

Growing a tree
- First we grow a tree using recursive binary splitting until some stopping criterion in met.
- regression uses RSS for this
- classicifcation grows using the Gini index
- Prune the tree: since the tree in the last time is probably too bushy and has high variance we trim it back using cross-complexity pruning. (use cross-validation to determine by how much)
We consider a sequence of trees indexed by a non-negative tuning parameter \(\alpha\). For every value of \(\alpha\) there corresponds a subtree, \(T \in T_0\), such that the following is minimized: \[\begin{equation} \sum_{m=1}^{|T|} \sum_{i:x_i \in R_m} (y_i - \hat y_{R_m})^2 + \alpha |T| \end{equation}\]
- Here \(|T|\) is the number of terminal nodes in the tree,
- \(R_m\) is the region corresponding to the \(m\)th terminal node
- \(\hat y_{R_m}\) is the mean of the training observations in \(R_m\).
Tuning Parameter
- When \(\alpha = 0\), then the subtree \(T\) will simply equal \(T_0\)
- However, as \(\alpha\) increases, there is a price to pay for having a tree with many terminal nodes, and so the penalized RSS will tend to be minimized for a smaller subtree.
- In that way, \(\alpha\) controls the trade-off between the subtree’s complexity and its fit to the training data.
- The tuning parameter \(\alpha\) is selected using cross-validation.

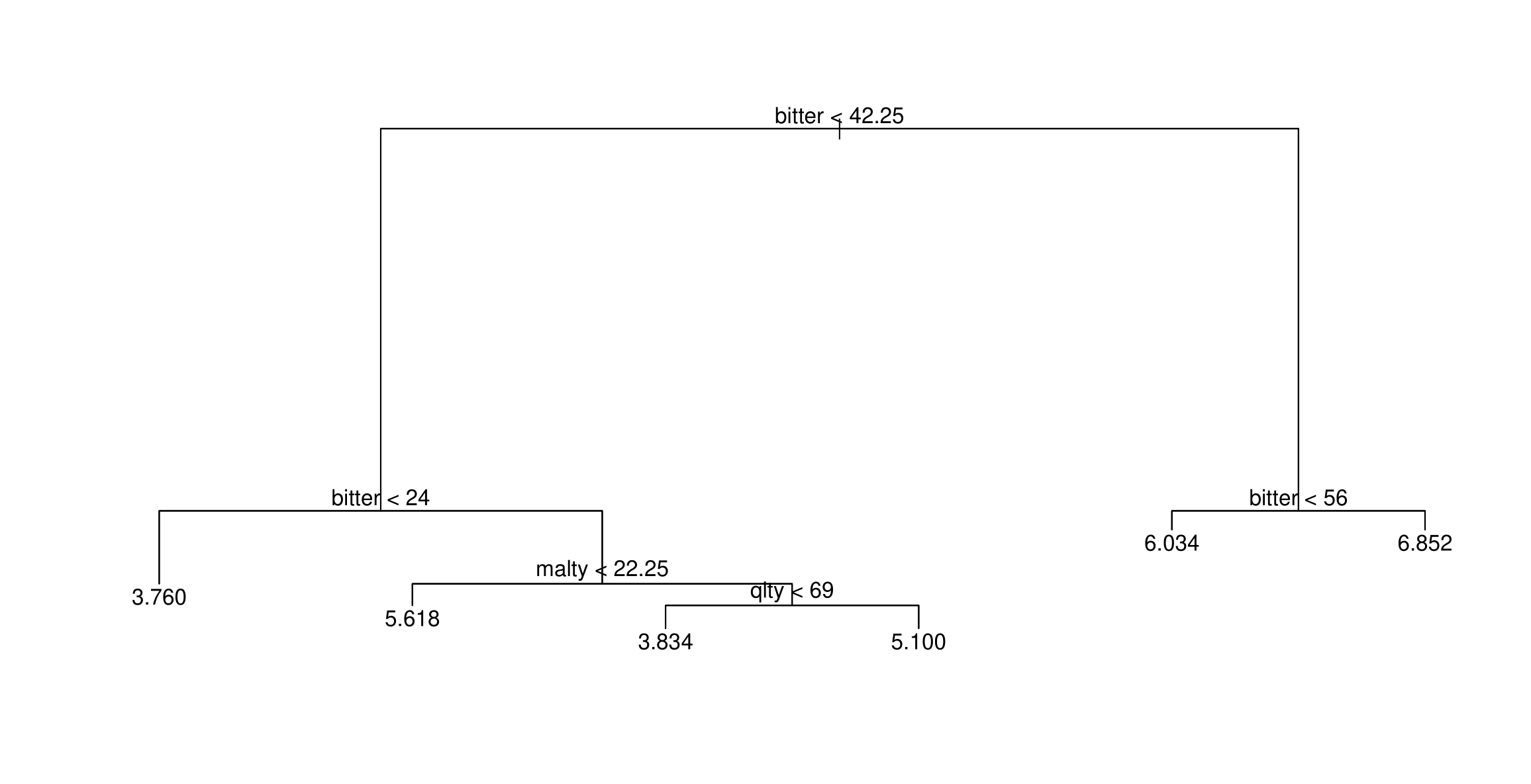
Trees have one aspect that prevents them from being ideal tool for predictive learning, namely inaccuracy. (ESL)
Lecture 14: Bagging and Random Forests

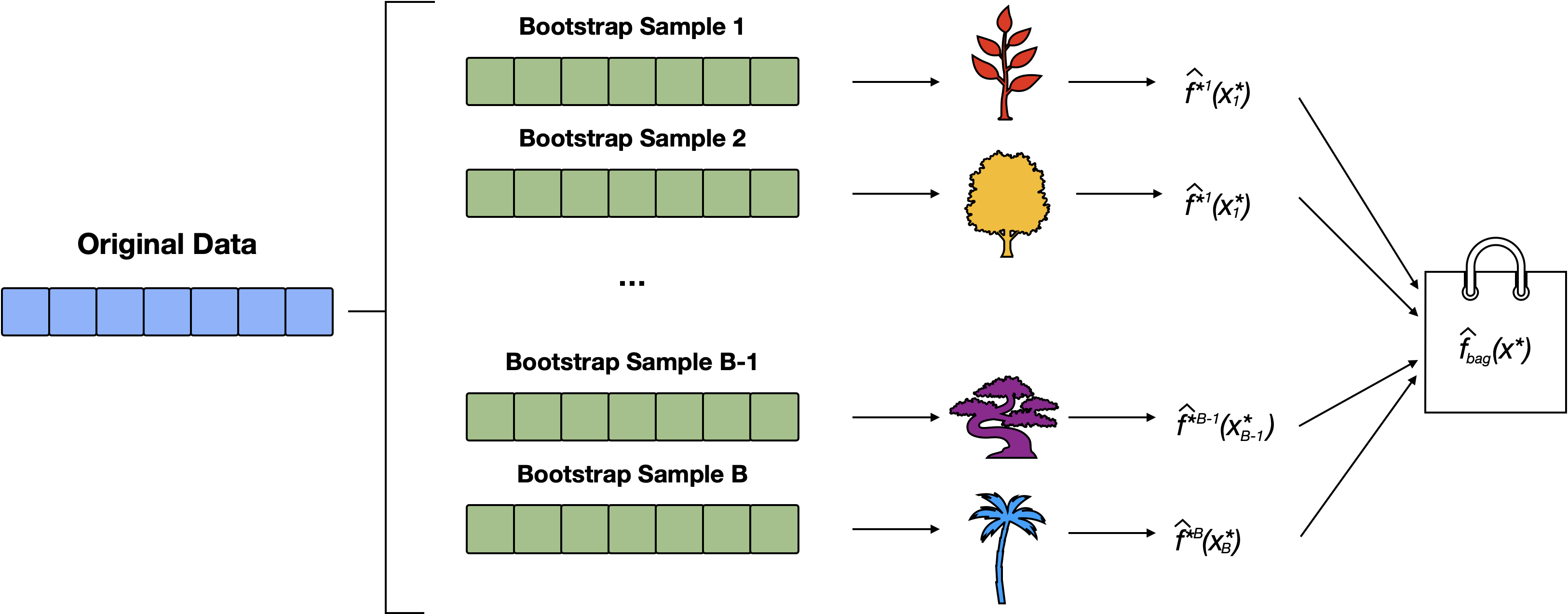
Bagging can be used to improve simple CART by reducing its variance.
Steps for Bagging
- Create \(B\) bootstrap samples1
- Fit a (bushy) tree to each bootstrap sample
- Each decision tree is used to make a prediction.
- Aggregate the \(B\) predictions:
- for regression predictions are typically averaged
- for classification we use a majority vote.
Random Forests uses a similar method to bagging, however, we only consider m predictors at each split
default for classification is \(m = \sqrt{p}\)
default for regression is \(m = p/3\)
Steps for RF
- Create \(B\) bootstrap samples
- Fit a tree to each bootstrap sample, HOWEVER, when creating nodes in each decision tree, only a random subset of \(m\) features is considered for splitting at each node.
- Each decision tree is used to make a prediction.
- Aggregate the \(B\) predictions.
This modification reduces the variance even more than bagging which tend to improve performance.
Both are fit with the same function in R:
randomForest(formula, data=NULL, mtry, importance = FALSE)when
mtry= \(p\) we get bagging and whenmtry\(< p\) we get random forests. Note that the default values are different for classification (\(\sqrt(p)\) where \(p\) is number of predictors) and regression (\(p/3\))when
importance = TRUEwe get variable importance computations (don’t worry about how they are calculated)
Model assessment
- The out-of-bag (OOB) samples (approximately 1/3 of the data) are used to create an cross-validation like estimate of error
- These are outputted by the
randomForest()function
Pros/cons
Pro:
- this approach prevents the risk of overfitting
- we get OOB error estimates “for free”
- makes trees a competive ML algorithm
Con:
- we’ve lost all interpretability
- takes longer to run
Lecture 15: Boosting
I have decided to remove this from the testable materials for the midterm.
Footnotes
in logistic regression, increasing \(X\) by one unit changes the log odds by \(\beta_1\)↩︎
where one class has significantly more instances than the other class or classes.↩︎
AKA city-block distance, L1 distance, \(\ell_1\) norm, or Taxicab geometry↩︎
\(B\) is usually quite is large (1000, 5000 are standard amounts)↩︎
\(B\) is usually quite is large (1000, 5000 are standard amounts)↩︎
where \(n\) is equal to the number of samples in your original data set↩︎
AKA the sample standard deviation
sd()of the \(\hat\alpha^{*i}\)s!↩︎