Data 311: Machine Learning
Lecture 6: Logistic Regression
University of British Columbia Okanagan
Introduction
Lately we’ve been focused on the regression problem–that is, the supervised machine learning problem when our response variable is a continuous quantity.
Machine learning algorithms that aim at predicting a categorical (AKA qualitative) response are referred to as classification techniques.
In this section, rather than trying to predict a continuous responses, we will be trying to classify observations in to categories (AKA classes)

Why not Linear Regression?
Consider coding categories as numeric, say we had recorded eye colour:
\[ Y = \begin{cases} 1 & \text{Green}\\ 2 & \text{Blue} \\ 3 & \text{Brown} \end{cases} \]
Suppose we fed this into a linear model.
Question: What would a prediction of 1.5 signify?
Class levels
Numeric codings for a response fed into a continuous model assumes a natural order, or hierarchy.
For multi-class categorical data, this is simply inappropriate.
- But what about a binary classification problem?
\[ Y = \begin{cases} 1 & \text{Green}\\ 0 & \text{Not green} \\ \end{cases} \]
Can we use this in a linear model such that a predicted value of 0.5 suggests a 50-50 chance that a person’s eye colour is green?
Example: body
Consider the
body(from the gclus package) data set from previous lecturesLet’s attempt to model the recorded
Genderas a linear function ofHeight.
Example: body
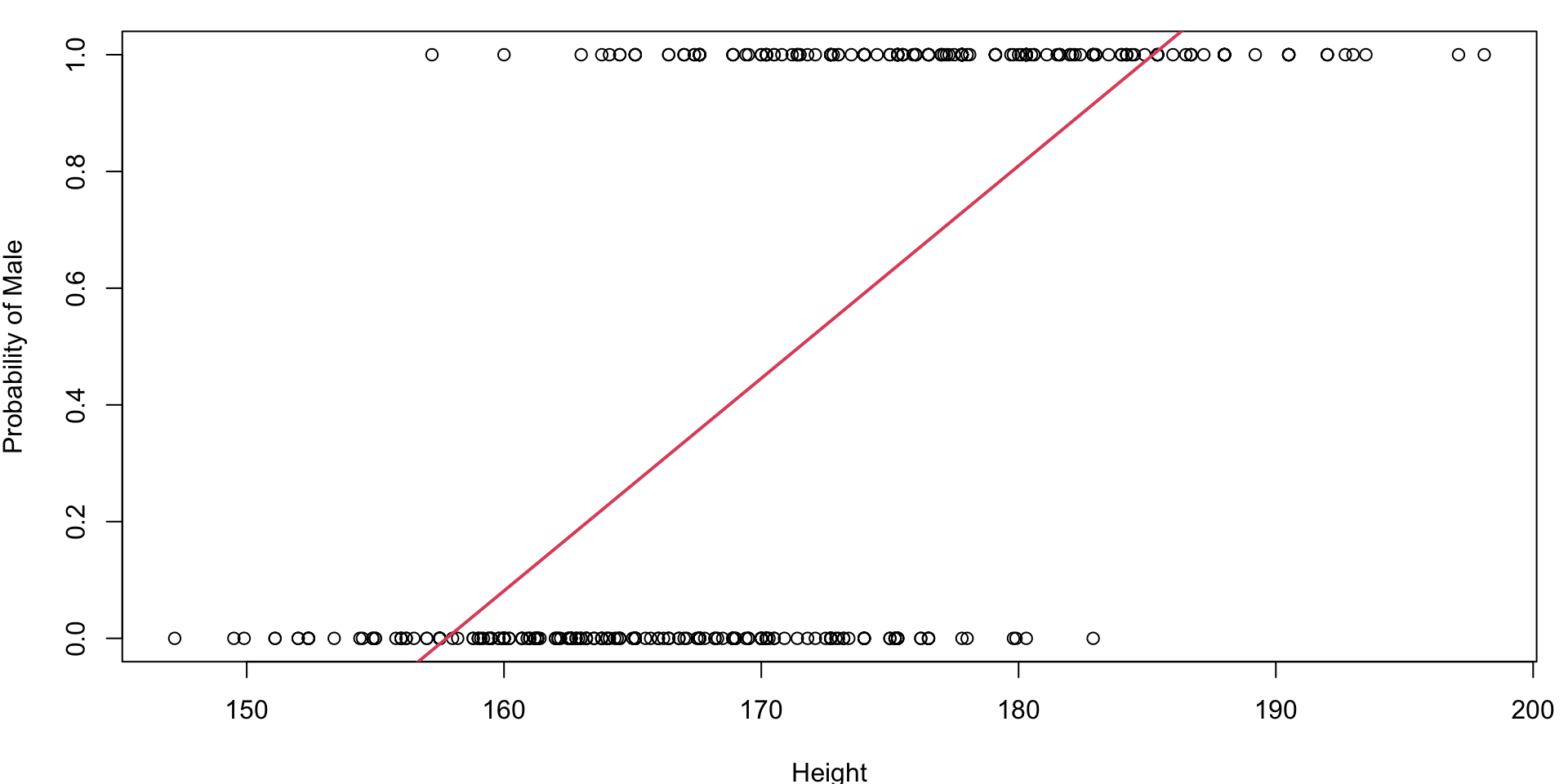
Multiple Linear Regression
Recall the linear regression model: \[\begin{equation} Y = \beta_0 + \beta_1 X_1 + \cdots + \beta_p X_p + \epsilon \end{equation}\]
- \(Y\) is the random, quantitative response variable
- \(X_j\) is the \(j^{th}\) random predictor variable
- \(\beta_0\) is the intercept
- \(\beta_j\) from \(j = 1, \dots p\) are the regression coefficients
- \(\epsilon\) is the error term
Fitted line
Same Plot (zoomed out)
Generalized Linear Model
Rather than modeling continuous \(Y \in (-\infty, \infty)\) using: \[\begin{equation} Y = \underbrace{\beta_0 + \beta_1 X_1 + \cdots + \beta_p X_p}_{\eta} \end{equation}\]
A GLM consists of three components:
- Systematic component
- Random component
- Link function
Systematic Component
The systematic component represents the linear combination of the independent variables.
It is typically expressed as \[\begin{equation} \eta = \beta_0 + \beta_1 x_1 + \dots \beta_p x_p \end{equation}\] where
\(\eta\) is the linear predictor,
\(\beta_0, \beta_1, \dots \beta_p\) are the coefficients, and
\(x_1, x_2, \dots, x_p\) are the predictors.
Random Component
We assume that \(y_1, \dots, y_n\) are samples of independent random variables \(Y_1, \dots, Y_n\) respectively.
The random component specifies the probability distribution \(f(y_i; \theta_i)\) of the response variable.
For GLM’s the probability distributions are assumed to arise from the Exponential family
Exponential family
The exponential family includes a large class of probability distributions e.g. normal, binomial, Poisson and gamma distributions, among others.
-
The mean, of the distribution depends on the independent variables, \(X\), through:
\[\begin{align*} \operatorname {E}[Y_i] &= g^{-1}(\eta)\\ \operatorname \eta &= g({E}[Y_i]) \end{align*}\]where \(g(\cdot)\) is the link function
Special cases of GLMs
Classical regression assumes:
- \(Y_i \sim \text{Normal}(\mu_i, \sigma^2)\)
- \(E[Y_i] = \mu_i = g^{-1}(\eta)\)
Logistic regression assumes
- \(Y_i \sim \text{Bernoulli}(\pi_i)\)
- \(E[Y_i] = \pi_i = g^{-1}(\eta)\)
We will studying binary (where \(Y\) is a binary random variable with two possible classes) but this can be extended to \(>2\) classes.
Link function
- The link function is a transformation that relates the mean of the response variable to the linear predictor.
If \(\eta = g(\mu_i) = \beta_0 + \beta_1 x_1 + \dots \beta_p x_p\) the link function is called the identity link and we get classical regression.
If \(\eta = g(\pi_i) = \ln \left( \frac{\pi_i}{1-\pi_i} \right)\) we get the logit link function and get logistic regression.
Logit Function
More generally if \(p\) is a probability, then
Plotted log-odds

If \(p = 0.5 \implies\) the odds \(\left(\frac{p}{1-p}\right)\) are even and the log-odds is zero.
Negative (resp. positive) logits represent probabilities below (resp. above) one half.
Activation Function
The link function \(g(\cdot)\) is an invertible function that transforms \(\operatorname {E}[Y_i]\) to \(\eta\).
However, in the machine learning community, we are often first introduced to the inverse of the link function \(g^{-1}()\).
This is sometimes called the activation function (or inverse link function.
For logistic regression, the activation function \(g^{-1}\) is the standard1 logisitic function (aka sigmoid function).
Relationship between Link and Activation Function
Link function \[\begin{align} g(\pi_i) &= \eta\\ \text{logit } \pi_i &= \eta\\ \ln \left( \dfrac{\pi_i}{1 - \pi_i} \right) &= \eta\\ \end{align}\]
Activation function \[\begin{align} \pi_i &= g^{-1}(\eta)\\ &= \dfrac{1}{1 + e^{-\eta}}\\ &= \dfrac{1}{1+ e^{-(\beta_0 + \beta_1 X_1 + \cdots \beta_p X_p)}} \end{align}\]
Logistic Function
\[\begin{align} g(\pi_i) &= \eta \\ \ln \left( \frac{\pi_i}{1-\pi_i} \right) &= \beta_0 + \beta_1 X_1 + \cdots \beta_p X_p \\ \frac{\pi_i}{1-\pi_i} &= \exp\{\beta_0 + \beta_1 X_1 + \cdots \beta_p X_p\} \\ \implies \pi_i &= \dfrac{\exp\{\beta_0 + \beta_1 X_1 + \cdots + \beta_p X_p\}}{1+ \exp\{\beta_0 + \beta_1 X_1 + \cdots \beta_p X_p\}}\\ &= \dfrac{1}{1+ e^{-(\beta_0 + \beta_1 X_1 + \cdots \beta_p X_p)}} = g^{-1}(\eta) = \dfrac{1}{1 + e^{-\eta}}\\ \end{align}\]This allows us to map the linear predictor \(\eta\) to probabilities (i.e. \(\pi_i\) values from 0 to 1)
Plotted Logistic Function
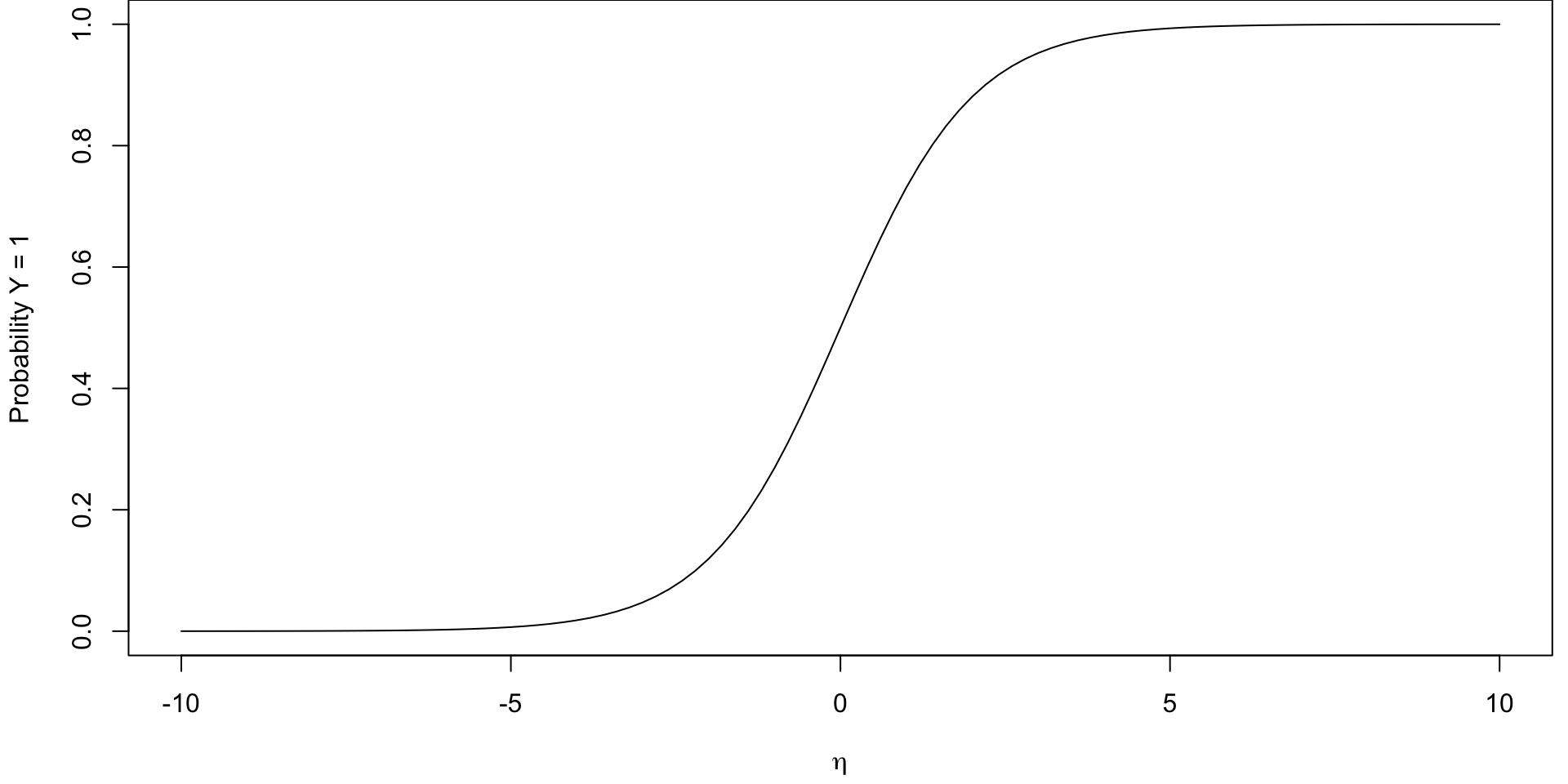
It is a S-shaped curve (sigmoid curve) that allows us to go back from logits to probabilities.
From Line to S-curve
Instead of trying to fit a line to this data, let’s try to fit something more like this S-curve so that \(0 \leq \pi_i \leq 1\)
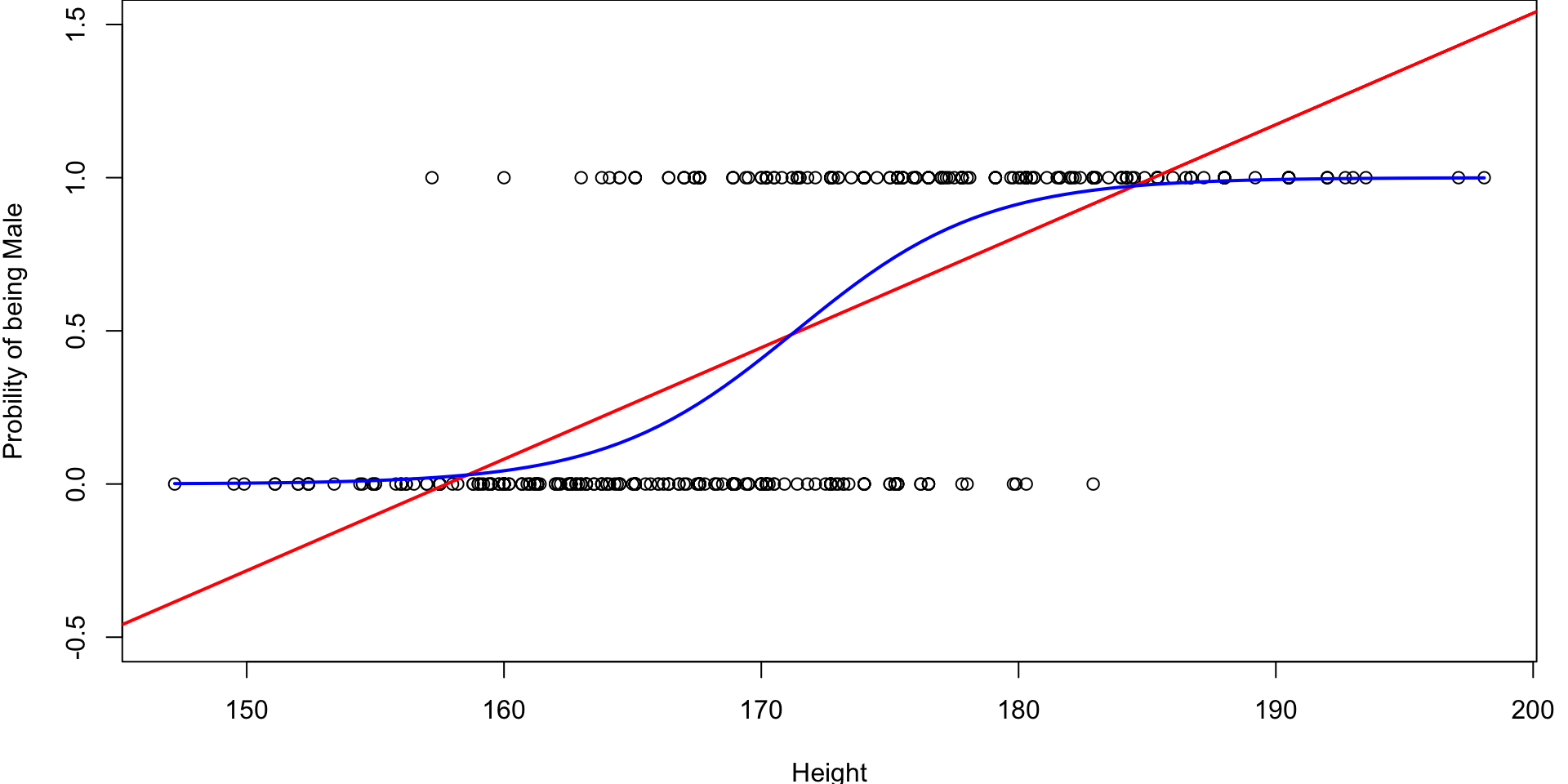
Logistic Regression
Logistic Regression:
\[ \text{log odds} = \beta_0 + \beta_1 X_1 + \dots + \beta_p X_p \]
Assumptions
\[ \begin{align} Y_i &\sim \text{Bernoulli}(\pi_i)\\ g({E} [Y_i]) &= \beta_0 + \sum_{j=1}^p\beta_j X_j = \eta \\ g^{-1}( \eta) &= \dfrac{e^{\sum_{j=1}^p\beta_j X_j}}{1+ e^{\sum_{i=1}^p\beta_i X_i}} = \dfrac{1}{1+e^{-\sum_{j=1}^p\beta_j X_j}}\\ \end{align} \]
where \(g(\cdot)\) is the logit function.
Interpretation of Coefficients
Since probability \(\pi_i\) is non-linear as \(X\) varies1, we cannot interpret the coefficients as we did in regular regression.
The best we can do is talk about the direction:
Holding all other variables constant, if \(\beta_j\) is positive, then an increase in \(X_j\) is associated with an increase in \(\pi_i\)
Holding all other variables constant, if \(\beta_j\) is negative, then an increase in \(X_j\) is associated with a decrease in \(\pi_i\).
Estimation
Let \(G_1\) be the set of observations where \(Y=1\), let \(G_0\) be the set of observations where \(Y=0\), and let \(\boldsymbol{\beta}\) be the set of coefficients. The likelihood is given by:
\[ l(\boldsymbol{\beta}) = \prod_{i \in G_1 }P(Y=1\mid x_i)\prod_{h \in G_0 }(1-P(Y=1\mid x_h)) \]
To fit the parameters of these models, we use maximum likelihood 1 estimation.
From lm() to glm()
You can fit a logistic regression using the glm() function.
| Arguments | Description |
|---|---|
formula |
a symbolic description of the model to be fitted, eg Y~ X1 + X2
|
family |
a description of the error distribution and link function to be used in the model ?family
|
data |
data frame (usually your training set) |
glm families
To perform logistic regression with a binary outcome we need to set
family = "binomial"1.-
Other options (not be covered in this course) include:
family = "gaussian"same aslm()family = "poisson"for predicting countsfamily = multinomialfor \(>2\) classesfamily = binomial('probit')probit regression (a common alternative to logistic regression)
Fitted Logistics Regression Plot
We are using the Logistic Function with the fitted \(\hat \beta\) values to plot the fitted S-curve
# Fit the logistic regression model
simlog <- glm(factor(Gender) ~ Height , family="binomial")
# store the beta hats for easy referencing
betas <- coef(simlog)
# plot the data (renaming the y-axis)
plot(Gender~Height, ylab="Probility of being Male")
# plot the fitted curve p(x) = e^(b0+b2x)/(1 + e^(b0+b2x))
curve(
(exp(betas[1] + betas[2]*x))/(1+exp(betas[1] + betas[2]*x)),
add=TRUE, # superimposes onto scatterplot (otherwise new plot)
lwd=2, # line width (make the line a bit thicker)
col=2) # 2 = red Fitted Logistics Regression Plot

Summary Output
...
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -46.76328 4.00571 -11.67 <2e-16 ***
Height 0.27292 0.02339 11.67 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 702.52 on 506 degrees of freedom
Residual deviance: 389.59 on 505 degrees of freedom
AIC: 393.59
Number of Fisher Scoring iterations: 5
...Multiple Logistic Regression
As with linear regression we could just as easily add predictors.
# Fit the logistic regression model
mlr <- glm(factor(Gender) ~ Height + WaistG + BicepG, family="binomial")
summary(mlr)...
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -55.32670 5.51960 -10.024 < 2e-16 ***
Height 0.21534 0.02864 7.520 5.50e-14 ***
WaistG 0.05167 0.02891 1.787 0.0739 .
BicepG 0.46541 0.08265 5.631 1.79e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 702.52 on 506 degrees of freedom
Residual deviance: 232.90 on 503 degrees of freedom
AIC: 240.9
Number of Fisher Scoring iterations: 6
...Residual Deviance
- You will notice that we no longer have a \(R^2\) or adjusted R-square value as we did in the linear regression case.
- Deviance is a lack of fit measure (the smaller the better) that plays the role of RSS for a broader class of models.
- The residual deviance is a measure of the deviance that remains unexplained after fitting the logistic regression model.
Evaluation
Evaluating a logistic regression model involves assessing its performance and determining how well it fits the data and makes predictions.
In Lecture 3 we saw some metrics for assessing regression models
Logistic regression, however, deals with the classification problem
In the realm of classification there are different set of evaluation techniques and metrics are used.
Metrics for Classification
In addition to deviance there are other popular metrics that we may consider for the classification problem:
- error/misclassification rate
- accuracy
- precision
- recall
- specificity
- F1-score
These can all be computed from a so-called classification table (aka confusion matrix)
Validation of Predicted Values
With a fitted logistic regression model the predict() function (see ?predict.glm) we can either predict the categorical response or output a probability.

From Probability to Classification
For predicting classification we consider
\[ \max \{ P(Y=0 \mid x), P(Y=1 \mid x) \} \]
We can obtain these probabilities in R using the predict() function on the fitted model (see lab)
Since we are considering only binary, \(P(Y=y \mid x)=0.5\) defines a the boundary.
Question: Where is this boundary for the body example?
Plot of Probability Boundary
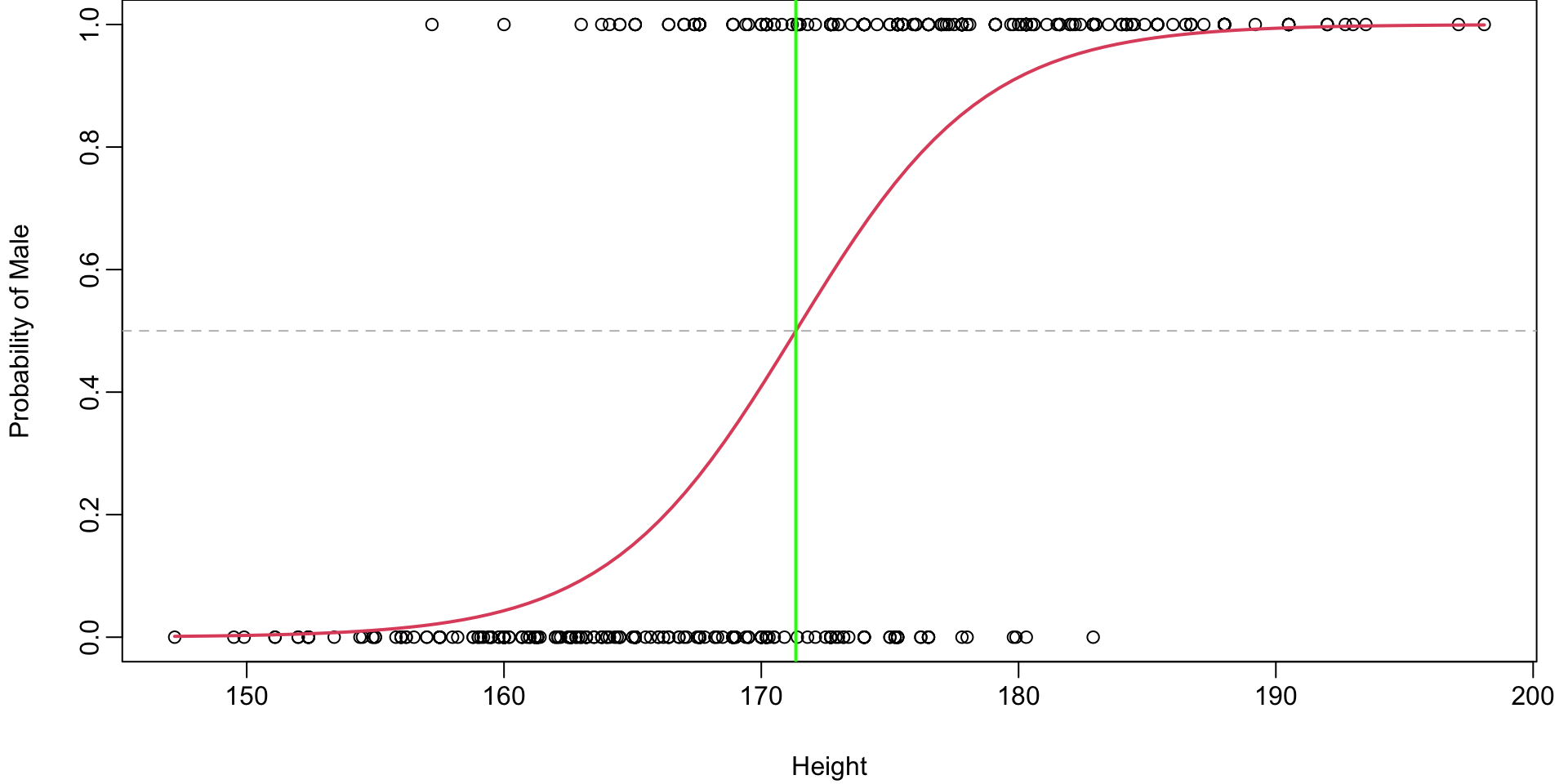
Height boundary
\[
\begin{align}
0.5
&=\dfrac{e^{\beta_0 + \beta_1 X}}{1+ e^{\beta_0 + \beta_1 X}}\\
\implies
\log \left(\frac{0.5}{0.5-0}\right) = \log \left(1\right) = 0 &= {\beta_0 + \beta_1 X} \\
\implies
X &= -\frac{\beta_0}{\beta_1}
\end{align}
\] So for Height \(> -\dfrac{ -46.7632791}{0.2729192} = 171.34\) we would predict Male.
Classification Table
For classification, we often summarize performance in a classification table (aka confusion matrix).
| predicted male | predicted female | |
|---|---|---|
| 1 - male | 216 | 44 |
| 0 - female | 45 | 202 |
In this case, the off-diagonals are errors (and diagonal are correctly classified obs.).
Error Rate
In words, the error rate (aka misclassification rate) calculates the proportion of misclassifications that are made by \(\hat f\):
\[\begin{align} \frac{1}{n} \sum_{i=1}^n I(y_i \neq \hat y_i) = \frac{\text{total misclassified}}{\text{total no. observations}} \end{align}\]and \(\hat y_i\) is the predicted class label for the \(i\)th observation \(x_i\) using \(\hat f\) and \[\begin{equation} I(y_i \neq \hat y_i)= \begin{cases} 1, y_i \neq \hat y_i\\ 0, \text{otherwise} \end{cases} \end{equation}\]
The error rate for this classification table is \[\begin{align} \dfrac{44+45}{216+44+45+202} = \dfrac{89}{507} = 0.1755424 \approx 17.6% \end{align}\]
Testing vs Training error rate
Akin to our earlier discussions with MSE for regression, we are generally interested with the error rate from the testing set rather than the training set.
That is, for a set of new observations \((x_0, y_0)\), a good classifier achieves the smallest test error on average: \[\begin{equation} E[I(y_0 \neq \hat y_0))], \end{equation}\] where \(\hat y_0\) is the predicted class label for \(x_0\) that uses \(\hat f\).
Comments
Your guess is as good as mine…(no more residuals to check)
No problem, bit of a change in notation and mathematics, but R can handle it just fine.
family = "multinomial"