Data 311: Machine Learning
Lecture 11: Cross Validation
University of British Columbia Okanagan
Introduction
In this lecture we’ll be looking at a resampling method called cross validation (or CV for short)
As it’s name suggests, this technique will be used for validating/accessing our model by providing an estimate of model performance on unseen data (i.e. not used in training).
Consequently, CV will be an indispensable tool for helping us choose between various modelling options.
Metrics for Model Assessement
For regression we aim to reduce the test MSE \[MSE = \frac{1}{N} \sum_{i=1}^N (y_i-\hat y_i)^2\]
For classification we may consider the test error rate \[\frac{1}{N} \sum_{i=1}^N I(y_i \neq \hat y_i)\]
Where \(1, \dots, N\) denotes “unseen” test observations that not used in the model-fitting process.
Recall
Ideally, we could fit our model on a sample of data, and then compare the predictions from the estimated model on a very large data set (not used in fitting) to estimate the MSE1.
If we only have one sample, we often split it into a training set (used to fit the model) and testing set (used to estimate the test MSE).
This train/test split approach is sometimes referred to validation approach.
Training vs Test-set Performance

Recall: training error rate often is quite different from the test error rate [Image source]
Validation Approach
Train/Test Split
By now, we have experience splitting our data set randomly into non-overlapping training and testing sets:
tr.ind
2, 3, 4, 6, 7, 8, 9, 10, 11, 14, …
te.ind
1, 5, 12, 13, 15, 18, 24, 31, …
In this case, we fit the model with samples with index tr.ind and estimate the MSE using samples with index te.ind
Validation Approach Schematic

A schematic display of the validation set approach. A set of n observations are randomly split into a training set (shown in red, containing observations 2,3,4,6,7,8,9,10,11,14,…, among others) and a validation set (shown in blue, containing observation 1, 5, 12, 13, 15, 18, 24, 31, 33, among others). The statistical learning method is fit on the training set, and its performance is evaluated on the validation set.
What are some issues you have encountered with this method?
ISLR Train Test Figure
jump forward to cross-validation approach

ILSR Fig 5.2 Compares linear vs. higher-order polynomial terms in Linear Regression Left: Validation error estimates for a single split into training (196 obs.) and validation (196 obs.) sets. Right: The validation method was repeated ten times, each time using a different random split of the observations into a training set and a validation set. This illustrates the variability in the estimated test MSE that results from this approach applied to the Auto data set.
Some concerns
High variability
- High variability in the estimate of the MSE using the test set.
- If we redo the process (new training set, new test set) the estimate can change considerably.
High bias
- This process will tend to overestimate the MSE by virtue of fitting the model to a smaller subset than we could have.
- reduced sample size will lead to poorer estimates
Resampling Methods
To combat this, it would be nice if we could have multiple testing and training sets.
-
While this is not a luxury we usually have, we can fudge it using resampling methods. This technique involves:
- repeatedly drawing samples from a training set
- refitting a model of interest on each sample
This will provide additional information about the fitted model that we would not otherwise get.
Motivation
We will discuss two of the most commonly used resampling methods: cross-validation (CV) and bootstrap
-
Today we focus on CV which can be used:
- to assess model performance (eg. estimate test MSE),
- perform model selection (eg. choose \(k\) for knn)
- model comparison (choosing between method 1 and 2)
- provide standard deviation and bias of our parameter estimates
Leave One Out Cross-Validation
Leave One Out Cross-Validation (LOOCV) is closely related to the validation approach but attempts to address some of that methods’ drawbacks
Like the validation set approach, LOOCV involves splitting the set of observations into two parts.
Unlike the validation approach, LOOCV systematically creates multiple training sets1 of size \(n - 1\) each having a validation (i.e. testing) set with a single observation \((x_i, y_i)\)
LOOCV approach
Full sample index (1, 2, … , 200) is systematically divided into:
Index
|
||
|---|---|---|
| CV Set | Training | Validation* |
| 1 | 2, 3, 4, ..., 200 | 1 |
| 2 | 1, 3, 4, ..., 200 | 2 |
| 3 | 1, 2, 4, ..., 200 | 3 |
| ... | ... | ... |
| 200 | 1, 2, 3, ..., 199 | 200 |
LOOCV Schematic

A schematic display of LOOCV. A set of n data points is repeatedly split into a training set (shown in red) containing all but one observation, and a validation set that contains only that observation (shown in blue). The test error is then estimated by averaging the n resulting MSE’s. The first training set contains all but observation 1, the second training set contains all but observation 2, and so forth.
LOOCV Steps
Steps:
Create \(n\) training/validation sets, then for each set:
Fit the model to the training set
Use the fitted model from 2. to predict the responses for the observation in the validation set.
Record the validation set error.
CV Estimation
If our response is quantitative, our validation set error can be assessed using the MSE.
For LOOCV the MSE corresponds to a single left out observation \(x_i\) will be defined as \(MSE_i = (y_i-\hat y_i)^2\) where \(\hat y_i\) is the prediction found from the \(i^{th}\) fitted model.
We can then estimate the MSE of the model using \[CV_{(n)} = \frac{1}{n} \sum_{i=1}^n MSE_i\]
LOOCV Steps Visualized
| Training Set | Prediction | Metric \(MSE_i\) |
|---|---|---|
| 2,3,4, …, 199, 200 | \(\hat y_1 = \hat f_{1}(x_1)\) | \((y_1 - \hat y_1)^2\) |
| 1,3,4, …, 199, 200 | \(\hat y_2 = \hat f_{2}(x_2)\) | \((y_2 - \hat y_2)^2\) |
| 1,2,4, …, 199, 200 | \(\hat y_3 = \hat f_{3}(x_3)\) | \((y_3 - \hat y_3)^2\) |
| \(\vdots\) | \(\vdots\) | \(\vdots\) |
| 1,2,3, …, 198, 199 | \(\hat y_{200} = \hat f_{200}(x_{200})\) | \((y_{200} -\hat y_{200})^2\) |
Pros to LOOCV
Less bias: LOOCV uses almost all of the data to fit the model whereas the validation approach may only use 50% (which tends to lead to an overestimate of the test error).
Deterministic estimates: LOOCV always gives the same estimate because the partitions are not chosen randomly.
Very general: LOOCV1 can be used with any kind of predictive modeling; logistic regression, LDA, you name it!
Cons to LOOCV
Computationally demanding: LOOCV requires the model to be fitted \(n\) times which can be very costly if \(n\) is large and/or the model is slow to fit.
More variance: LOOCV estimates are highly correlated1 so recall: \[\begin{align} \text{Var}(X + Y) = \text{Var}(X) + \text{Var}(Y) + 2\cdot\text{Cov}(X,Y) \end{align}\]
\(k\)-Fold CV
We can consider other alternatives of cross validation instead of leave-one-out.
We can randomly subdivide the sample into \(k\) approximately1 equally-sized and non-overlapping sets
Each set (or “fold”) takes a turn as the validation set, with the remainder of the data used to train the model.
Typically we use 5- or 10-fold CV.
5-fold Visualization

A schematic display of 5-fold CV. A set of n = 20 observations is randomly split into five non-overlapping groups (or folds). Each of these folds takes a turn a validation set (shown in blue), and the remainder as a training set (shown in red). The test error is estimated by averaging the five resulting MSE estimates.

\(k\)-Fold split
Full Sample Index: 1, 2, … , 20, with \(k = 5\)
| Training Set | Validation* |
|---|---|
| 1, 2, 4, 5, 6, 8, 9, 11, 13, 14, 15, 16, 17, 18, 19, 20 | 3, 7, 10, 12 |
| 1, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 15, 16, 17, 18, 20 | 2, 4, 14, 19 |
| 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12, 14, 15, 17, 19, 20 | 5, 13, 16, 18 |
| 1, 2, 3, 4, 5, 7, 10, 11, 12, 13, 14, 16, 17, 18, 19, 20 | 6, 8, 9, 15 |
| 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 13, 14, 15, 16, 18, 19 | 1, 11, 17, 20 |
5-fold CV Calculations
| Validation Set \(V\) | Prediction | Metric \(MSE_j\) |
|---|---|---|
| \(V_1\) = 3, 7, 10, 12 | \(\hat f_1 (x_3, x_7, x_{10}, x_{12})\) | \(\frac{1}{4}\sum_{i\in V_1} (y_i - \hat y_i)^2\) |
| \(V_2\) = 2, 4, 14, 19 | \(\hat f_2 (x_2, x_4, x_{14}, x_{19})\) | \(\frac{1}{4}\sum_{i\in V_2} (y_i - \hat y_i)^2\) |
| \(V_3\) =5, 13, 16, 18 | \(\hat f_3 (x_5, x_{13}, x_{16}, x_{18})\) | \(\frac{1}{4}\sum_{i\in V_3} (y_i - \hat y_i)^2\) |
| \(V_4\) =6, 8, 9, 15 | \(\hat f_4 (x_6, x_8, x_{9}, x_{15})\) | \(\frac{1}{4}\sum_{i\in V_4} (y_i - \hat y_i)^2\) |
| \(V_5\) =1, 11, 17, 20 | \(\hat f_5 (x_1, x_{11}, x_{17}, x_{20})\) | \(\frac{1}{4}\sum_{i\in V_5} (y_i - \hat y_i)^2\) |
\(k\)-Fold Estimation
More generally, we can calculate the MSE for each validation set \(j\), for \(j = 1, \dots, k\) \[MSE_j = \frac{1}{n_j} \sum_{i \in j}(y_i-\hat y_i)^2\] where \(n_j\) is the number of observations in the \(j\)th validation set. A cross-validated estimate the MSE given by \[CV_{(k)} = \sum_{j=1}^K \frac{n_j}{n} MSE_j \]
ISLR Cross Validation Figure
jump back to validation approach

ILSR Fig 5.4 Cross-validation was used on the Auto data set in order to estimate the test error that results from predicting mpg using polynomial functions of horsepower. Left: The LOOCV error curve. Right: 10-fold CV was run nine separate times, each with a different random split of the data into ten parts. The figure shows the nine slightly different CV error curves. These are much less variable than the Validation Approach
\(k\)-fold vs. LOOCV
Pros of \(k\)-fold
- Less variance than LOOCV1
- More computationally feasible than LOOCV (\(k\) vs. \(n\) model fits)
- Often gives more accurate estimates of test error than LOOCV.
Cons of \(k\)-fold
- Slightly more bias than towards overestimating MSE in comparison to LOOCV
- Non deterministic estimate of the MSE (randomly spliting the data into \(k\) folds)
Bias-Variance trade-off


LOOCV will have lower bias than does \(k\)-fold CV with \(k < n\), but LOOCV has higher variance.
k-fold or LOOCV?
- k-fold or LOOCV
k-fold or LOOCV?
-
k-fold or LOOCV?1 What value of \(k\) is best?
The ISLR authors argue that for intermediate values of \(k\), say 5 or 10, \(k\)-fold CV provides a better estimate of test error rate than LOOCV2
In other words, the decrease in variance tends to outweigh the increase in bias when we chose 5/10-fold validation over LOOCV.
CV and Classification (LOO)
CV (either \(k\)-fold or LOO) can be applied easily in a classification context as well.
For instance, in LOOCV we can calculate cross-validated misclassification rates by replacing \(MSE_i\) with the validation misclassification rate (aka error rate \(\text{Err}_i\)) : \[ CV_{(n)} = \frac{1}{n} \sum_i^n \text{Err}_i = \frac{1}{n} \sum_i^n I(y_i \neq \hat y_i) \] where \(I(y_i \neq \hat y_i)\) is 1 if the left-out observation was misclassified using the \(i\)th model, and 0 otherwise.
CV and Classification (k-fold)
We divide the data into \(k\) roughly equal-sized parts \(C_1, C_2, \ldots, C_k\) where \(n_j\) is the number of observations in part \(j\): if \(n\) is a multiple of \(k\), then \(n_j = n/k\).
\[\begin{align} MC_j &= \frac{1}{n_j} \sum_{i \in j} I(y_i \neq \hat y_i) \\ CV_{(k)} &= \sum_{j=1}^k \frac{n_j}{n} MC_j \end{align}\]Model Selection
We have just seen how we could use these methods to to assess model performance.
Now let’s switch gears and see how these methods can be used for model selection.
Now we can use cross-validation (CV) to compare and select the best-performing model among several candidates.
In this case, we use CV to choose which value of
k1 for KNN performs the best in terms of generalization.
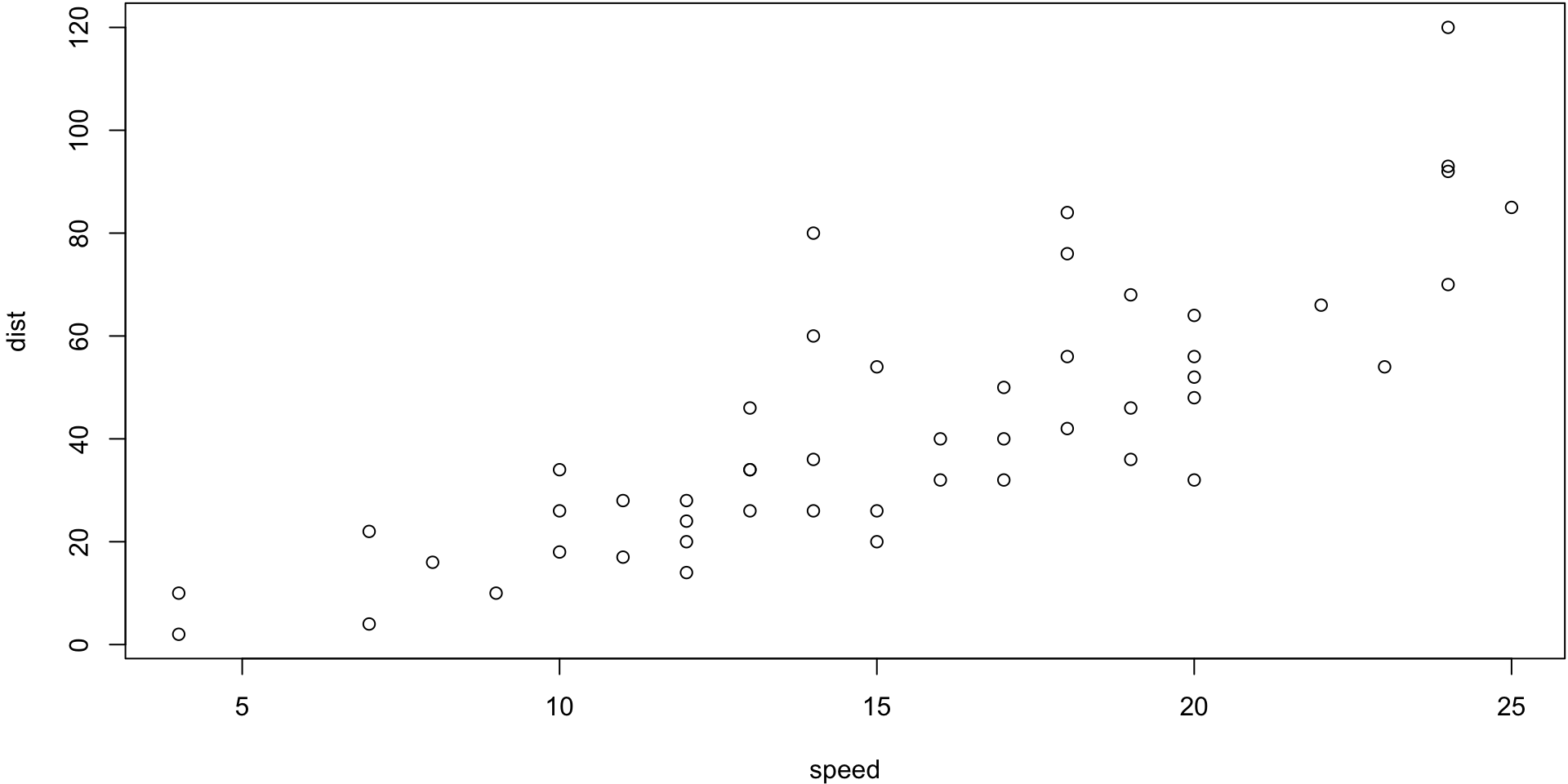
Cars Example
The cars data give the speed of 1920s cars (speed) and the distances taken to stop (dist).
KNN (from FNN package)
-
trainmatrix or data frame of training set cases. -
testmatrix or data frame of test set cases. A vector will be interpreted as a row vector for a single case. If not supplied, LOOCV will be done. -
yreponse of each observation in the training set. -
knumber of neighbours considered.
Cross-validated estimates
We fit all possible
kvalues (1 to 49 - the number of samples)If we don’t supply a test test,
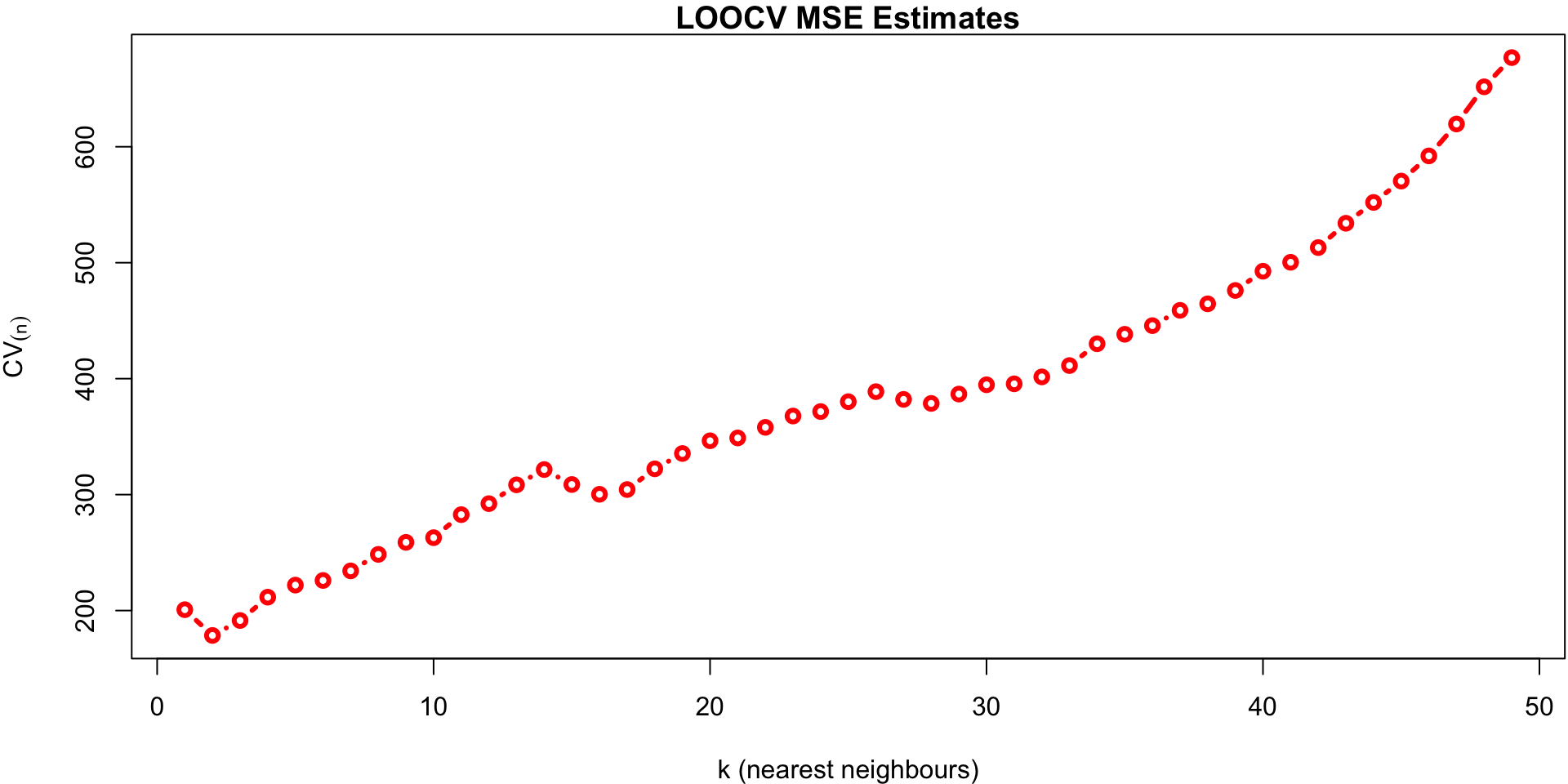
knn.regperforms LOOCVWe plot the cross-validated estimates of MSE across all values of
keach calculated using: \[CV_{(n)} = \frac{1}{n} \sum_{i=1}^n MSE_i\]
k vs MSE plot
library(FNN)
data(cars, package="datasets")
attach(cars)
kr <- list()
predmse <- NA
for(i in 1:49){
# LOOCV (default if no test set is supplied)
kr[[i]] <- knn.reg(speed, test=NULL, dist, k=i)
predmse[i] <- kr[[i]]$PRESS/nrow(cars) # LOOCV estimate for k = i
}
par(mar = c(4, 4.2, 1.1, 0.1))
plot(predmse, type="b", lwd=3, col="red",
ylab = expression(CV[(n)]),
xlab = "k (nearest neighbours)",
main ="LOOCV MSE Estimates")
k vs MSE plot
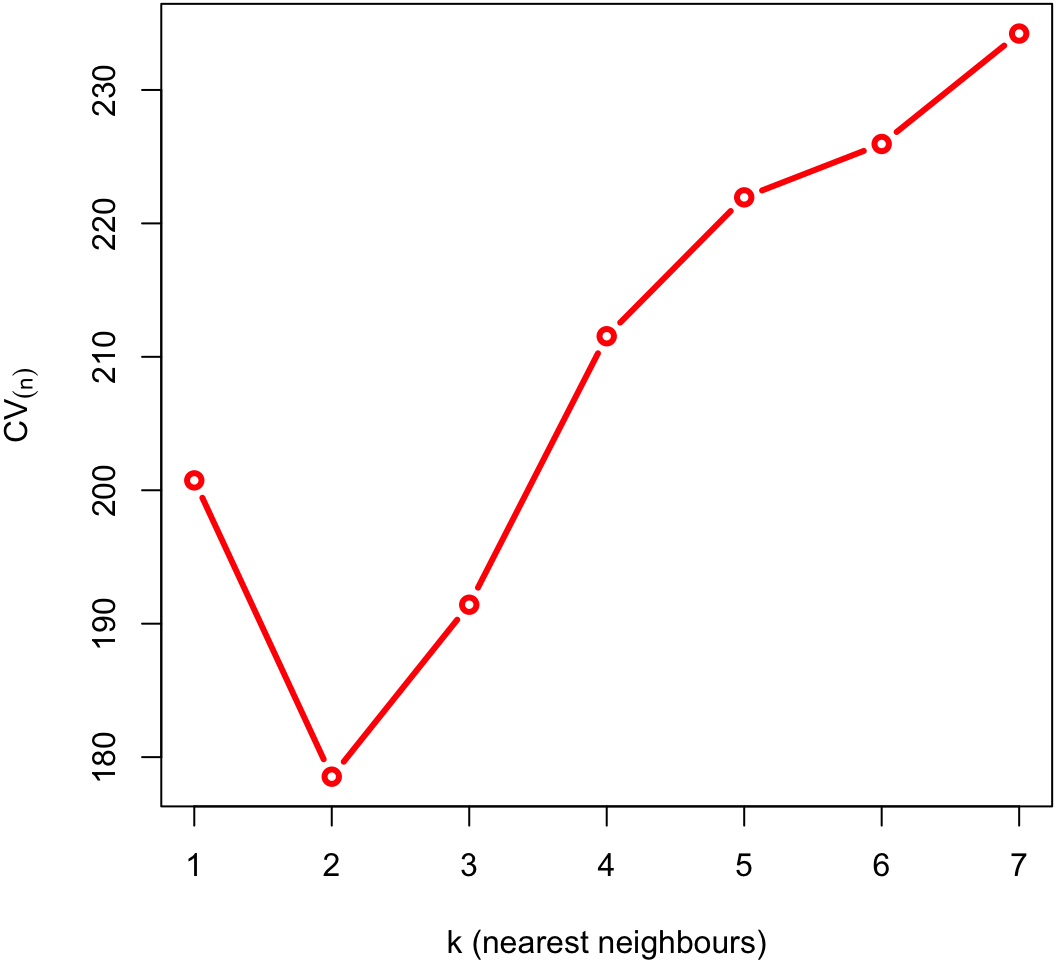
Zoomed in
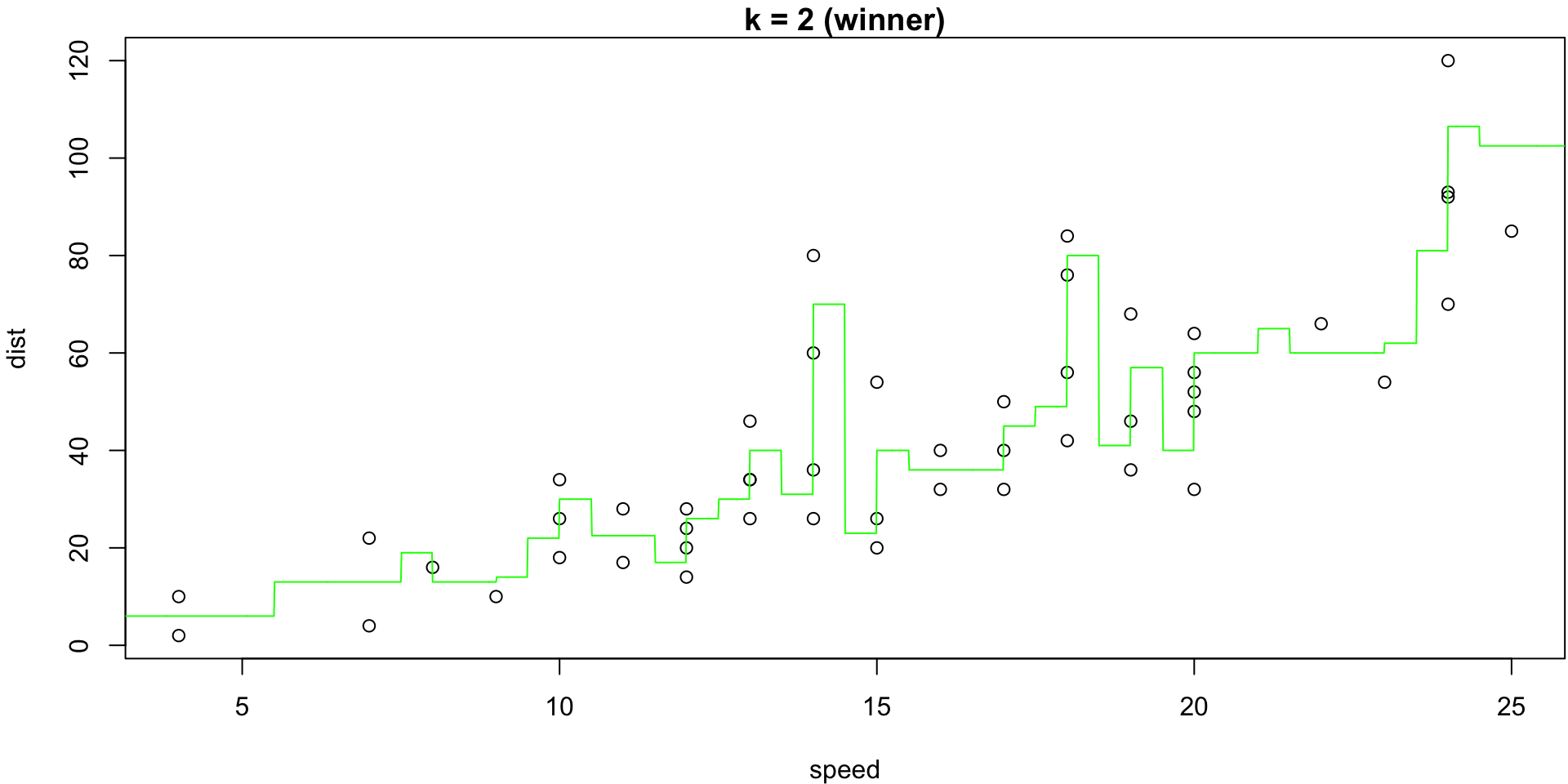
The minimum of that plot suggests that the best value of k is at 2.
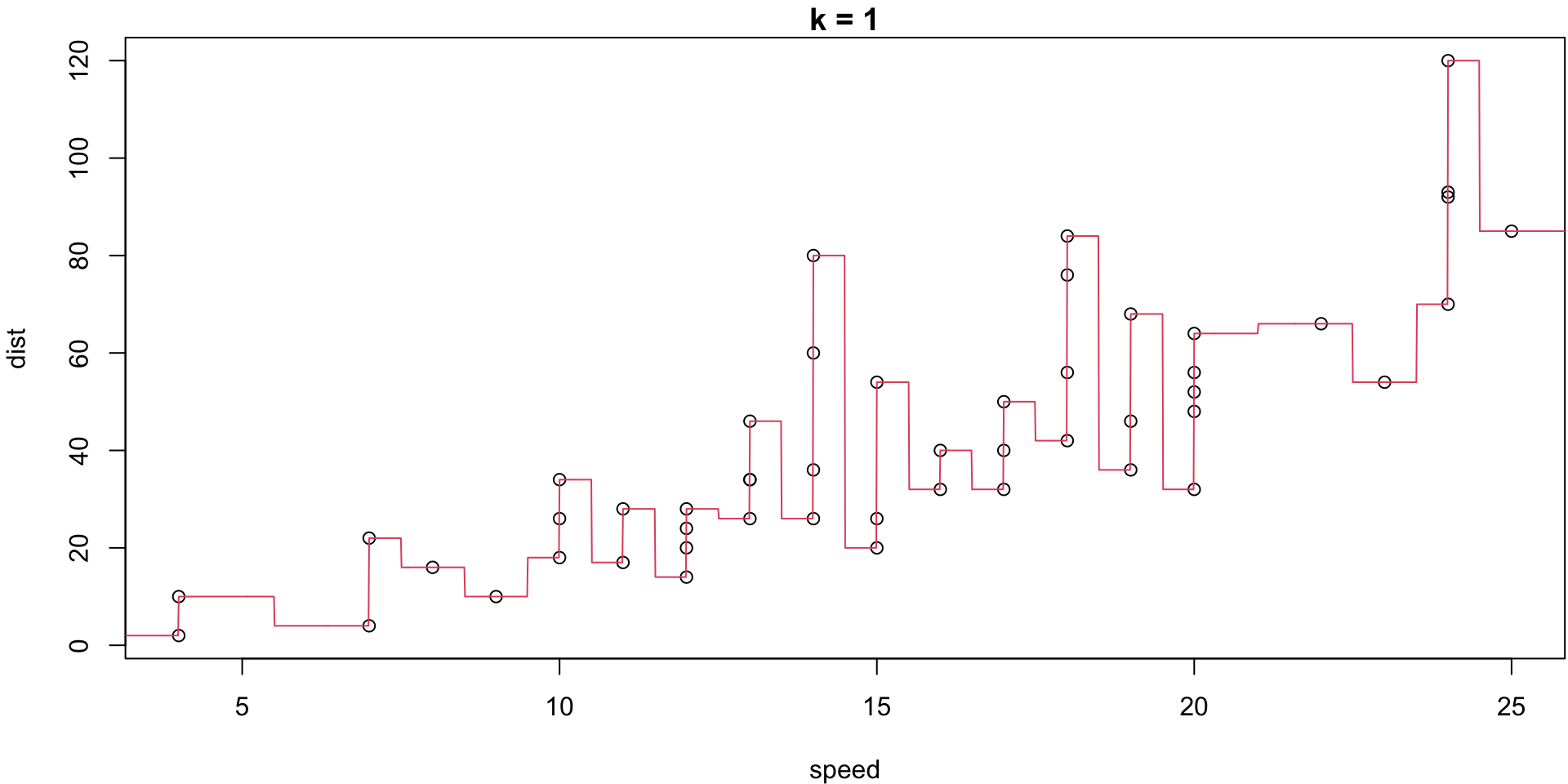
k = 1
k = 5

k = 20

k = 2 (winner)
Comparing Models
Not only can we select
kusing CV, but we can furthermore compare that best KNNreg model’s predictive performance with any other potential model.Since CV estimates the MSE, there’s no reason we cannot provide that estimate in the context of, say, simple linear regression…
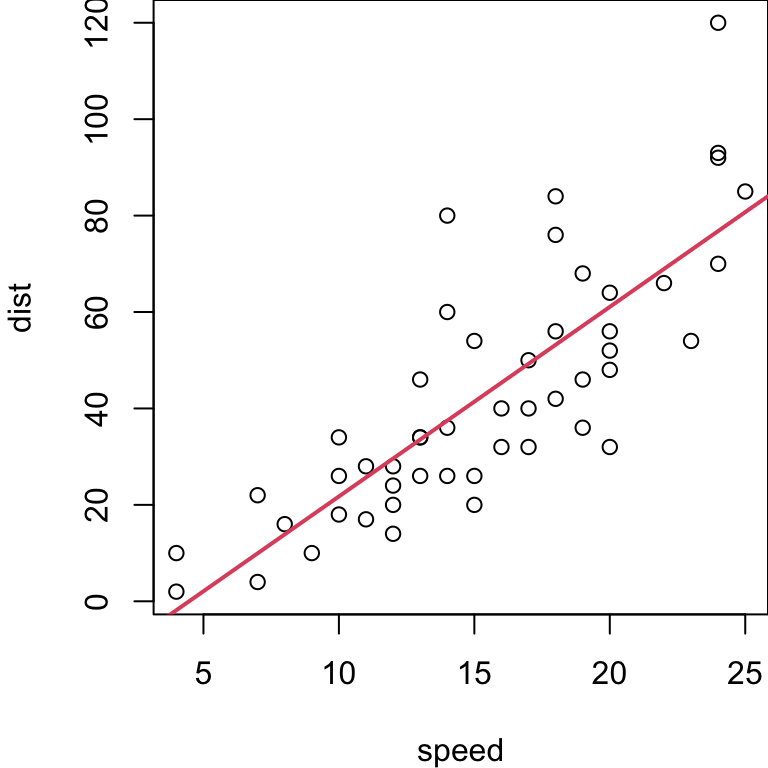
Cars example (regression)
Car data, simple linear regression on full sample
...
Call:
lm(formula = dist ~ speed)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -17.5791 6.7584 -2.601 0.0123 *
speed 3.9324 0.4155 9.464 1.49e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 15.38 on 48 degrees of freedom
Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438
F-statistic: 89.57 on 1 and 48 DF, p-value: 1.49e-12
...Regression MSE estimate
Under the inferential assumptions for SLR, we have a theoretical (unbiased) estimate of the test MSE via \[ \hat{\text{MSE}} = \frac{\text{RSS}}{n-2} = \frac{\sum (y_i - f(x_i))^2}{n-2}\]
For the
carsdata, that gives us: \(\frac{11353.52}{48}= 236.53\)In this case
k= 2 KNN model gets a CV MSE estimate of 178.54 hence is suggested as the better model.
LOOCV Regression Shortcut
While we could compare these estimates of MSE directly, we could go one step further and get the CV estimate for MSE by applying LOOCV to the linear model!
Notably, there is a mathematical shortcut for LOOCV with the least squares linear or polynomial regression that calculate \(CV_{(n)}\) using a single fit!
The details of this shortcut are discussed in Section 5.1.2 of the ILSR textbook (we won’t go over them here).
LOO Regression Animation
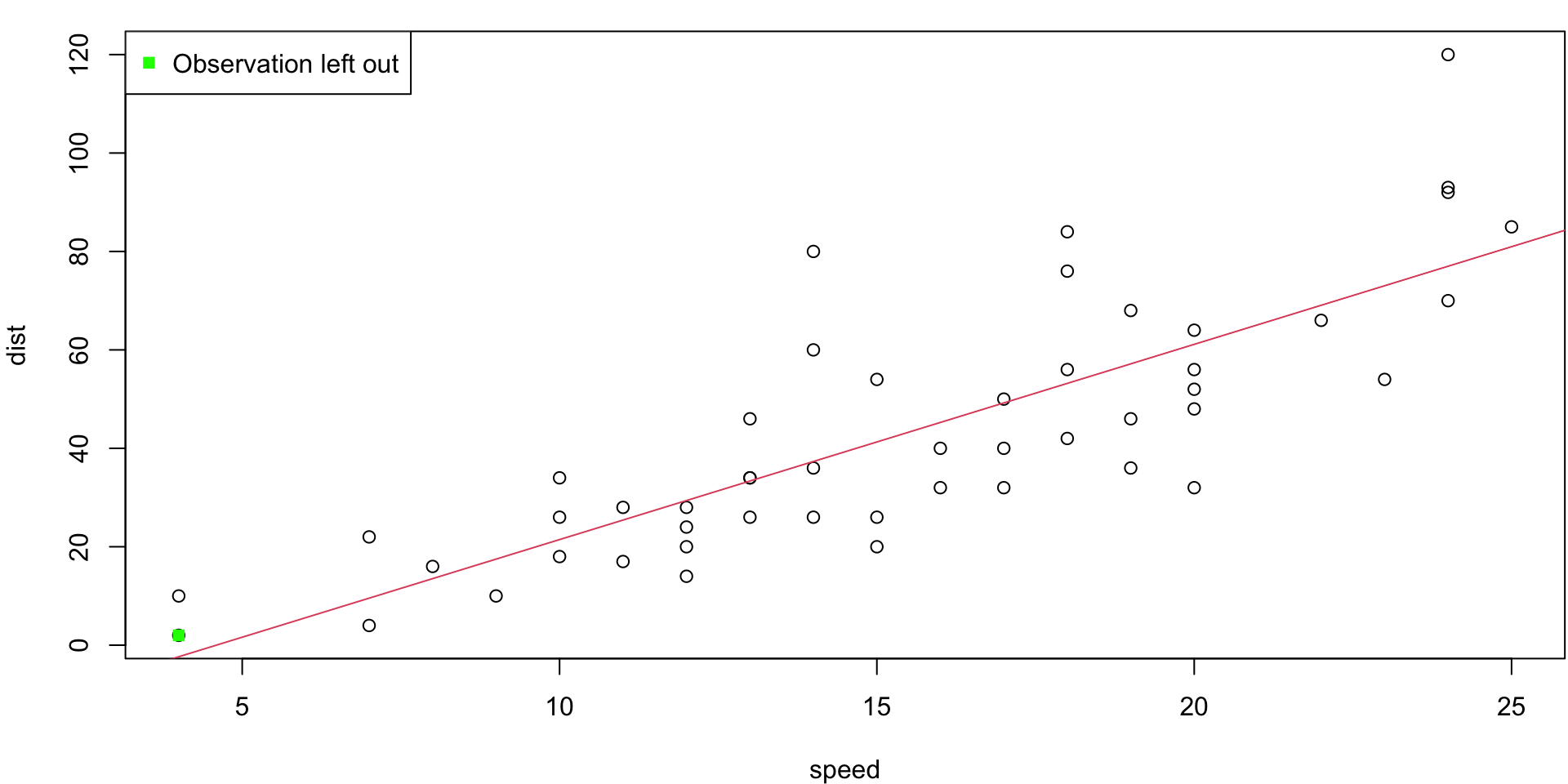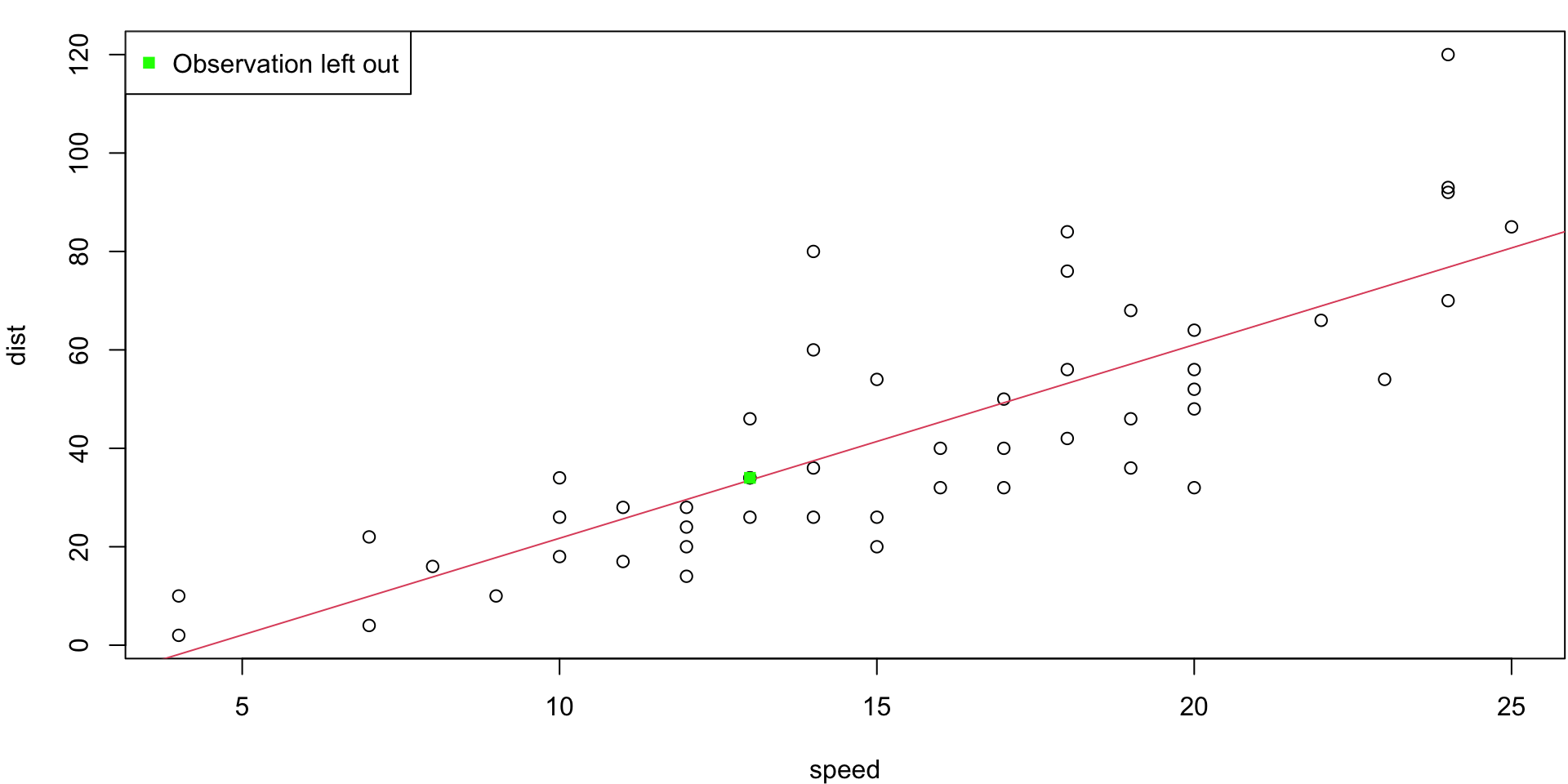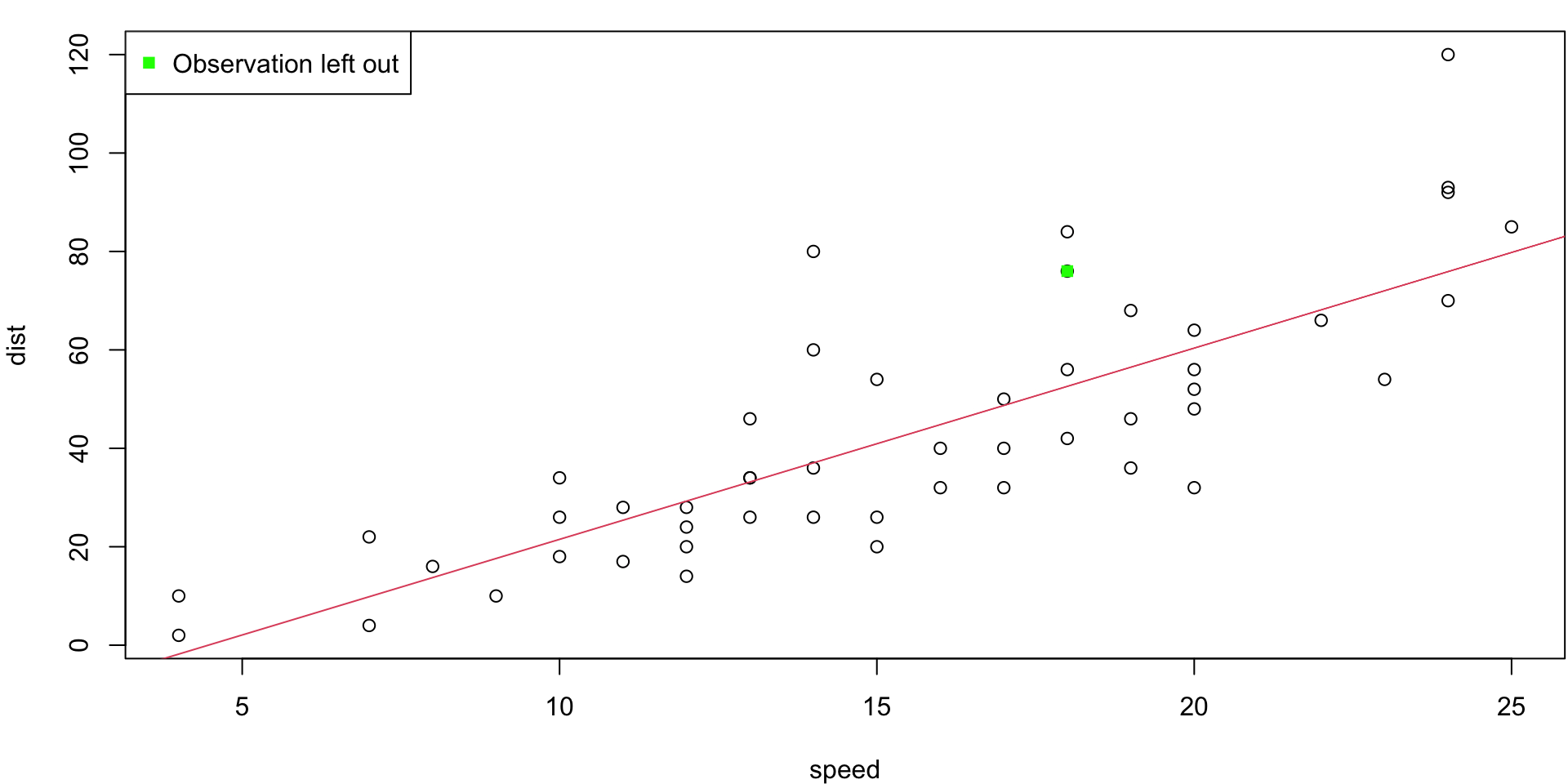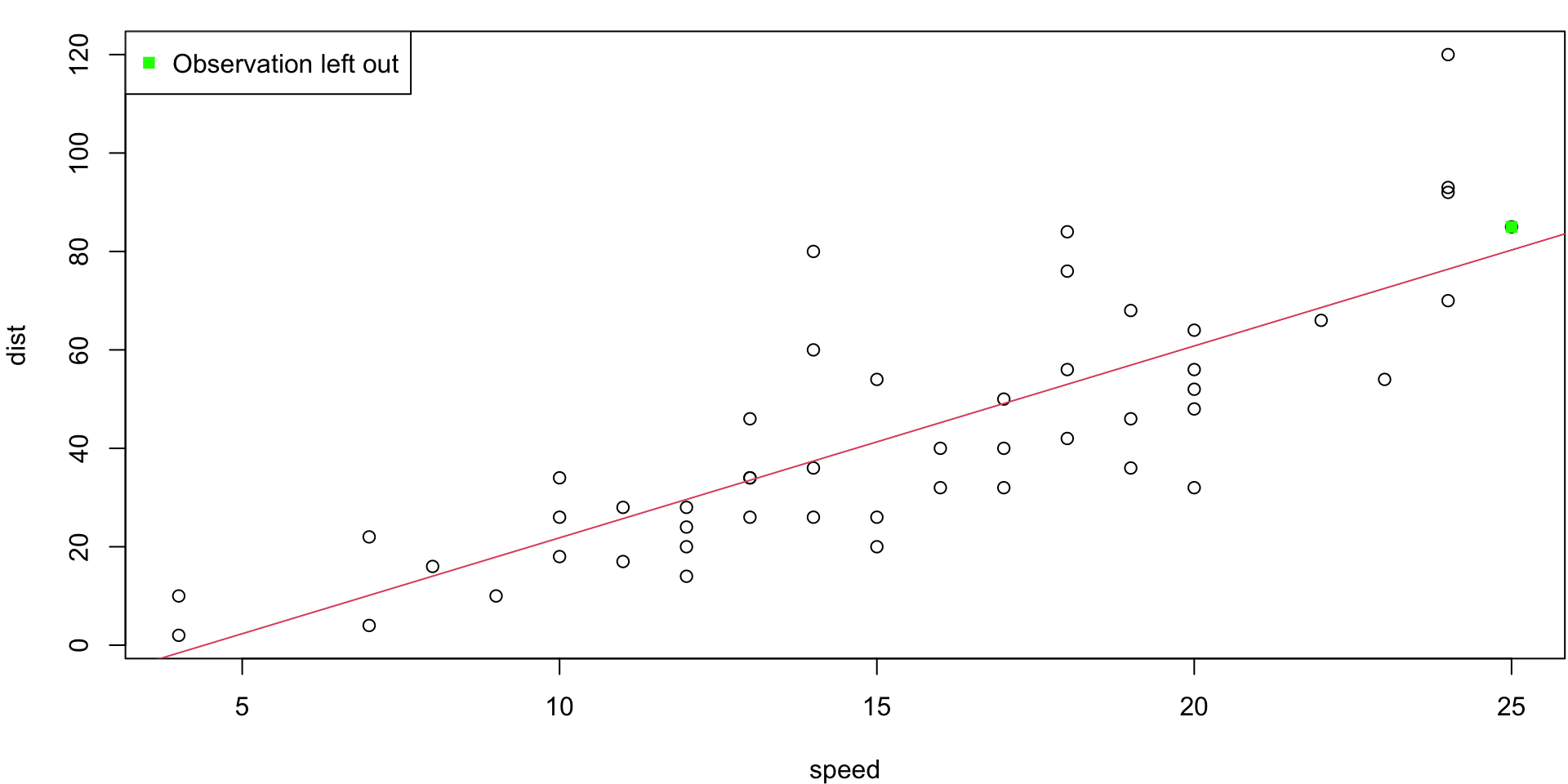
LOO regression fits
Plots the \(n\) (highly correlated) LOOCV regression fits produce a cross-validated estimates MSE = \(CV_{(n)} = 246.405416\)
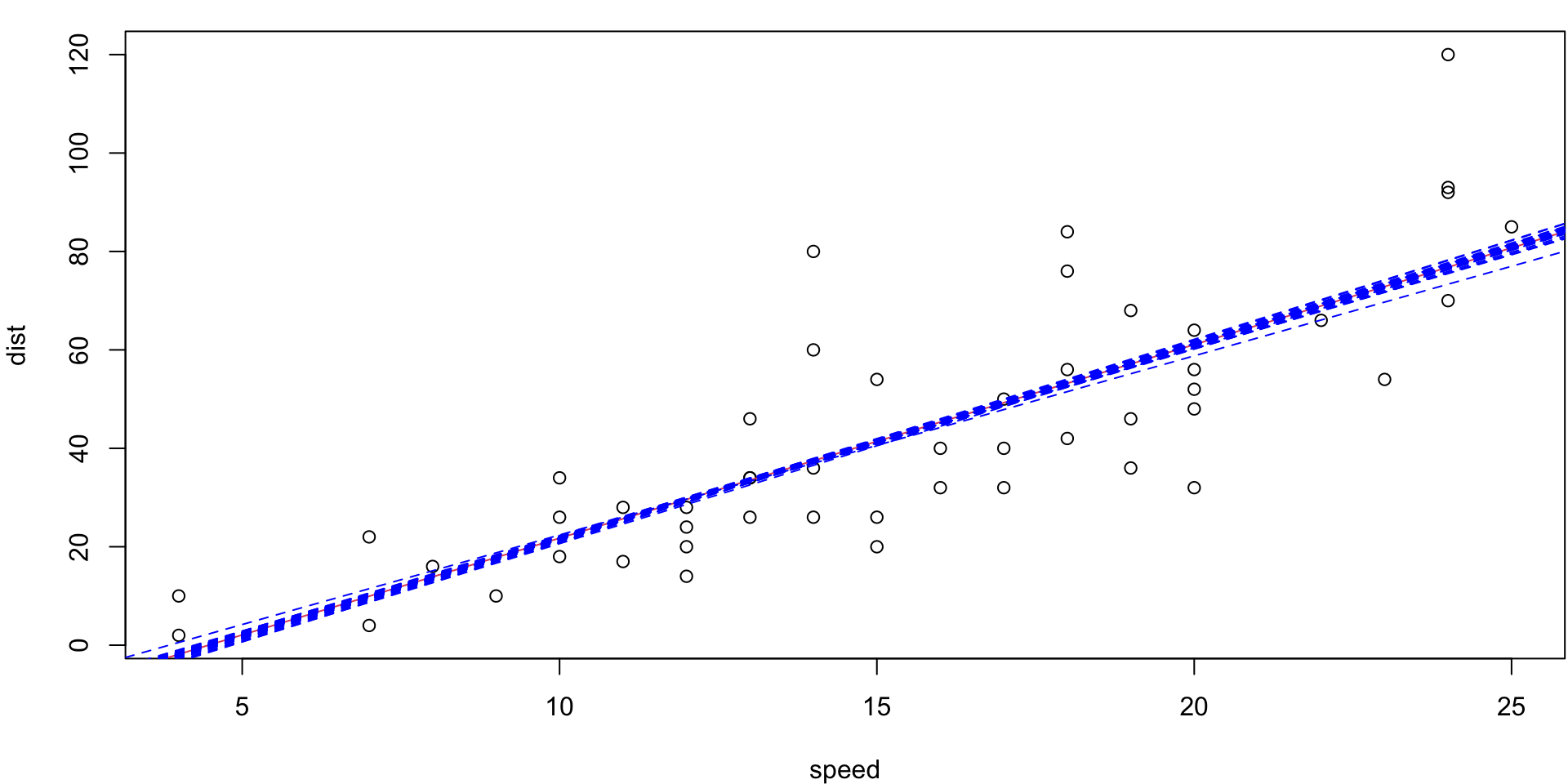
5-fold CV Animation
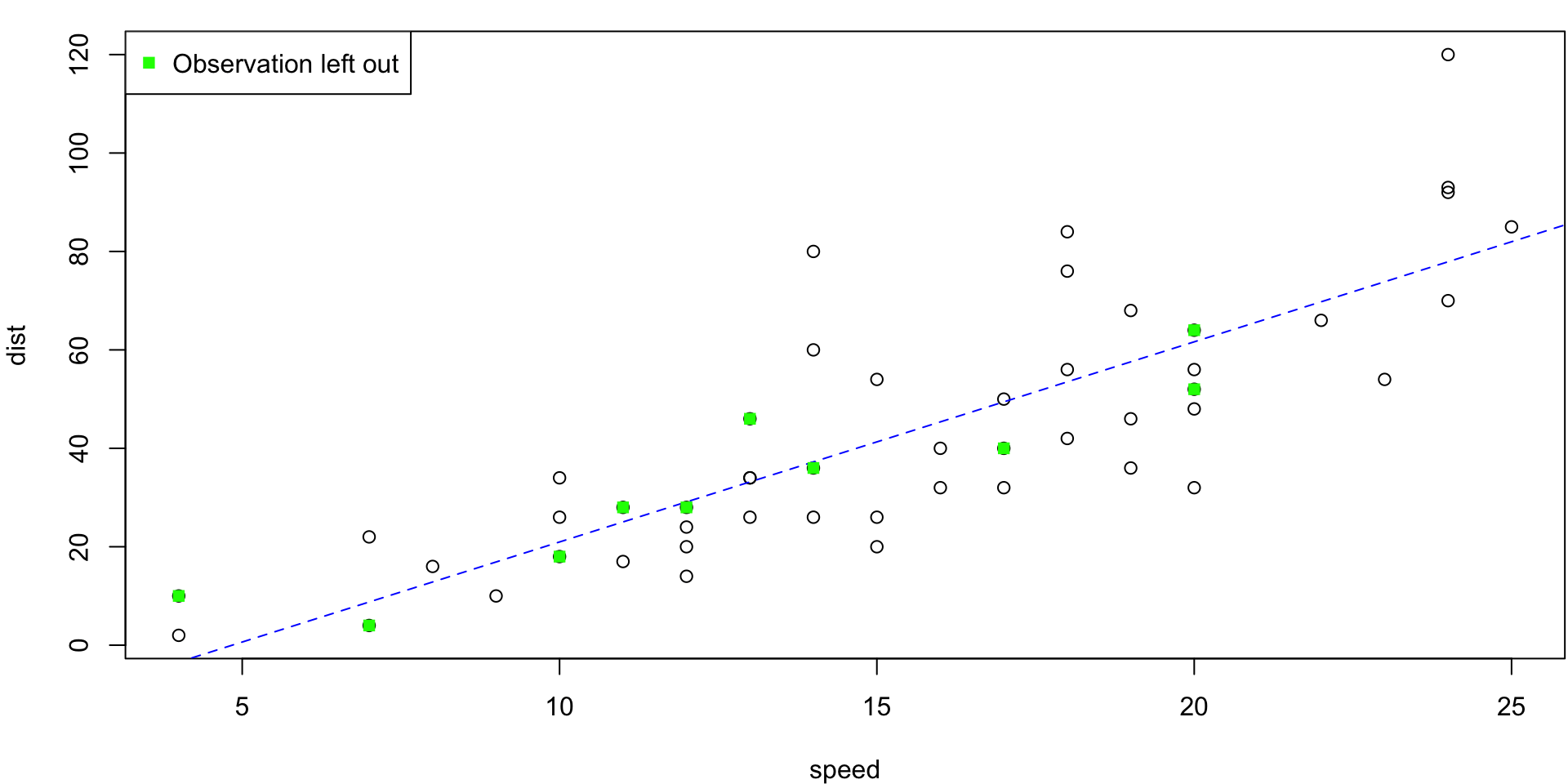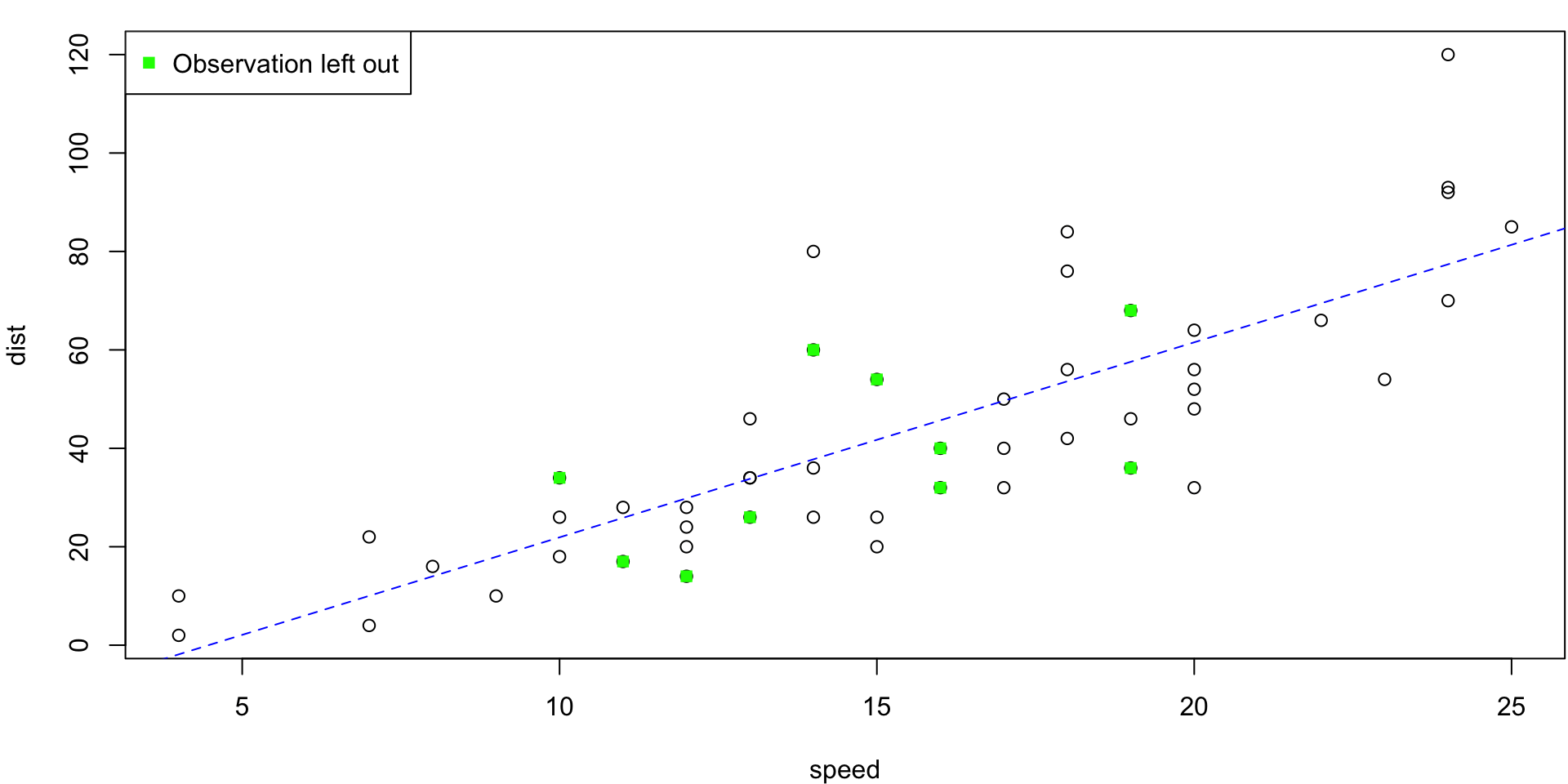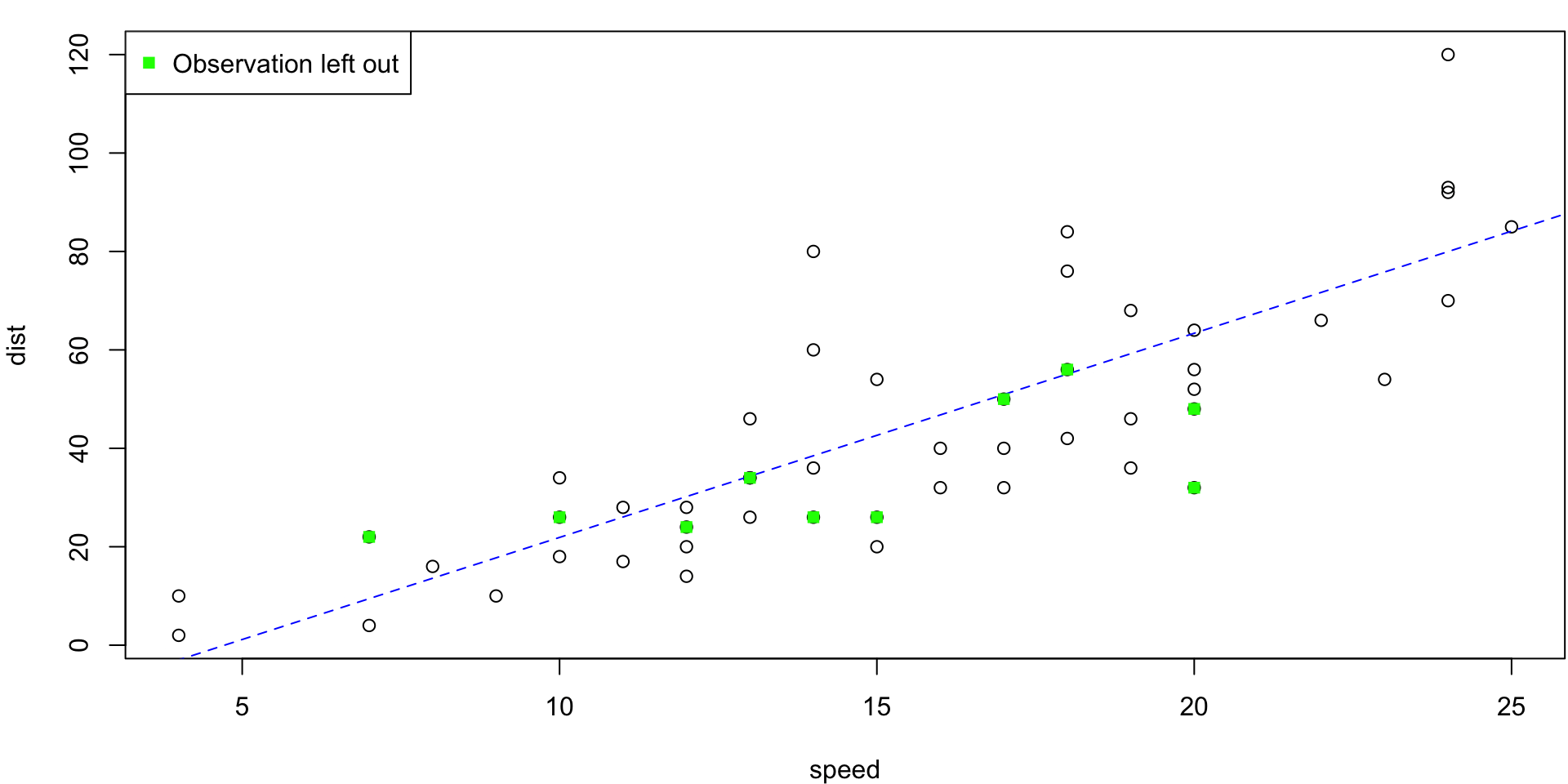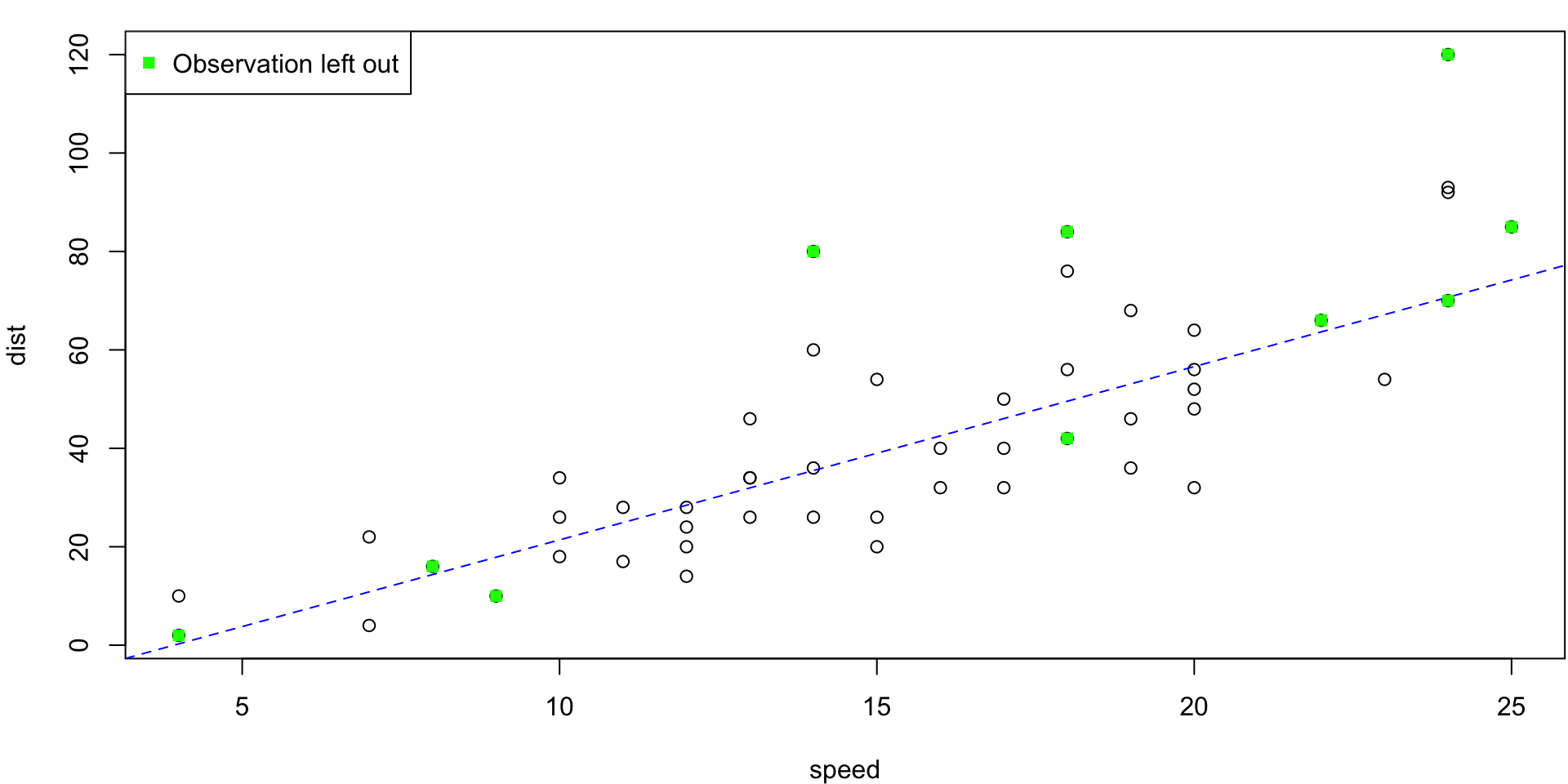
5-fold estimates MSE as \(CV_{(5)} = 254.0391118\)

10-fold CV Animation
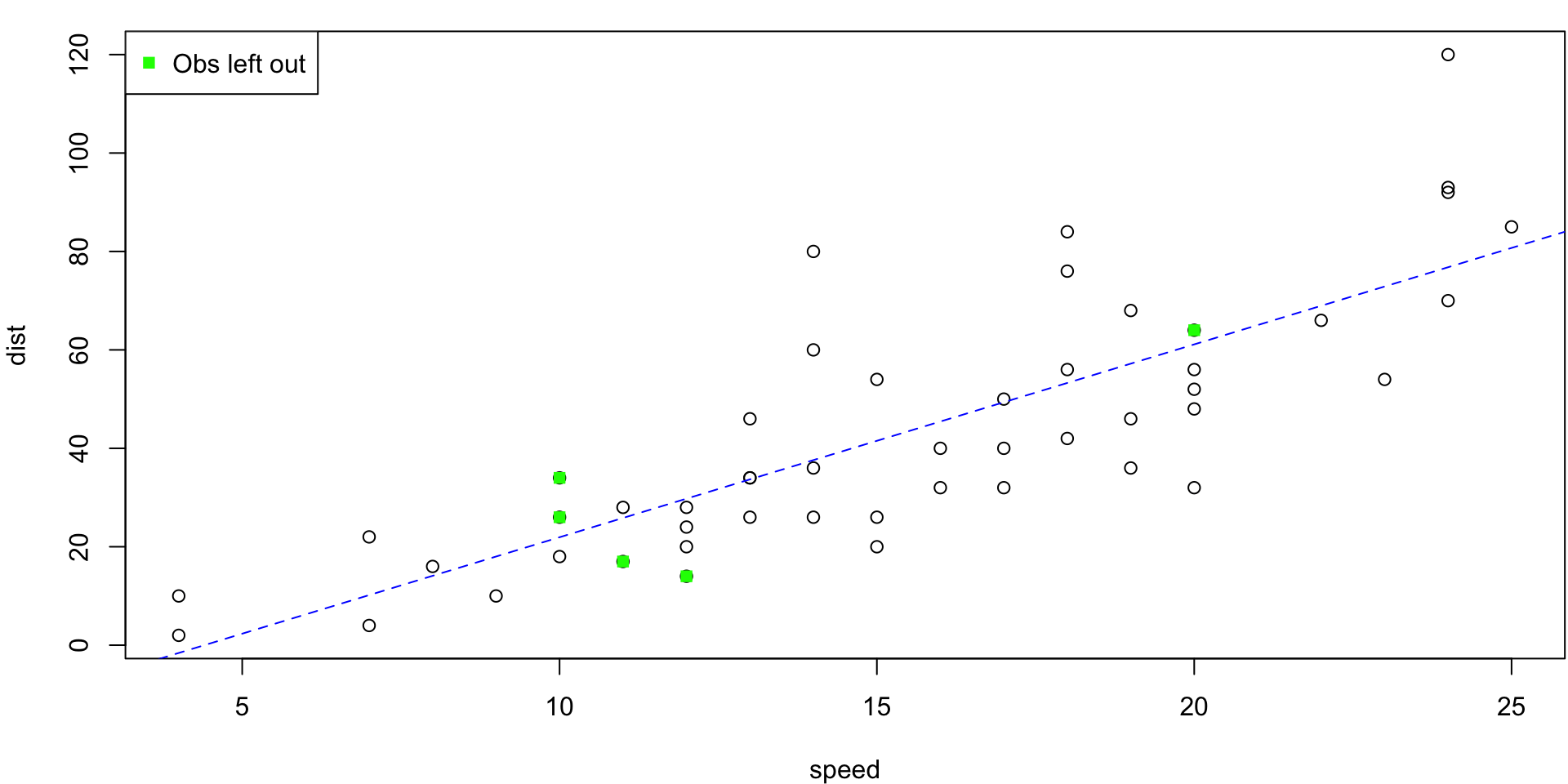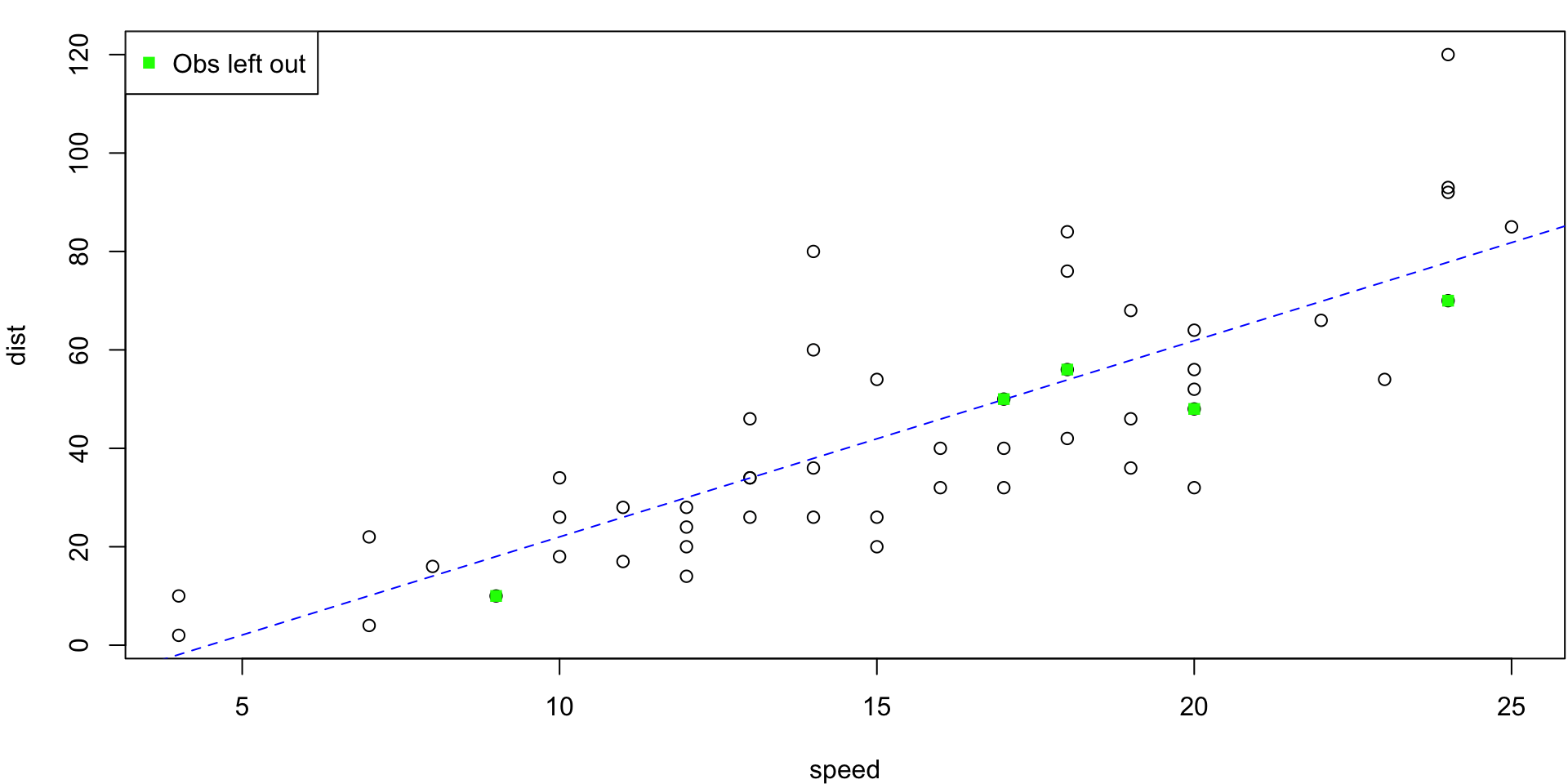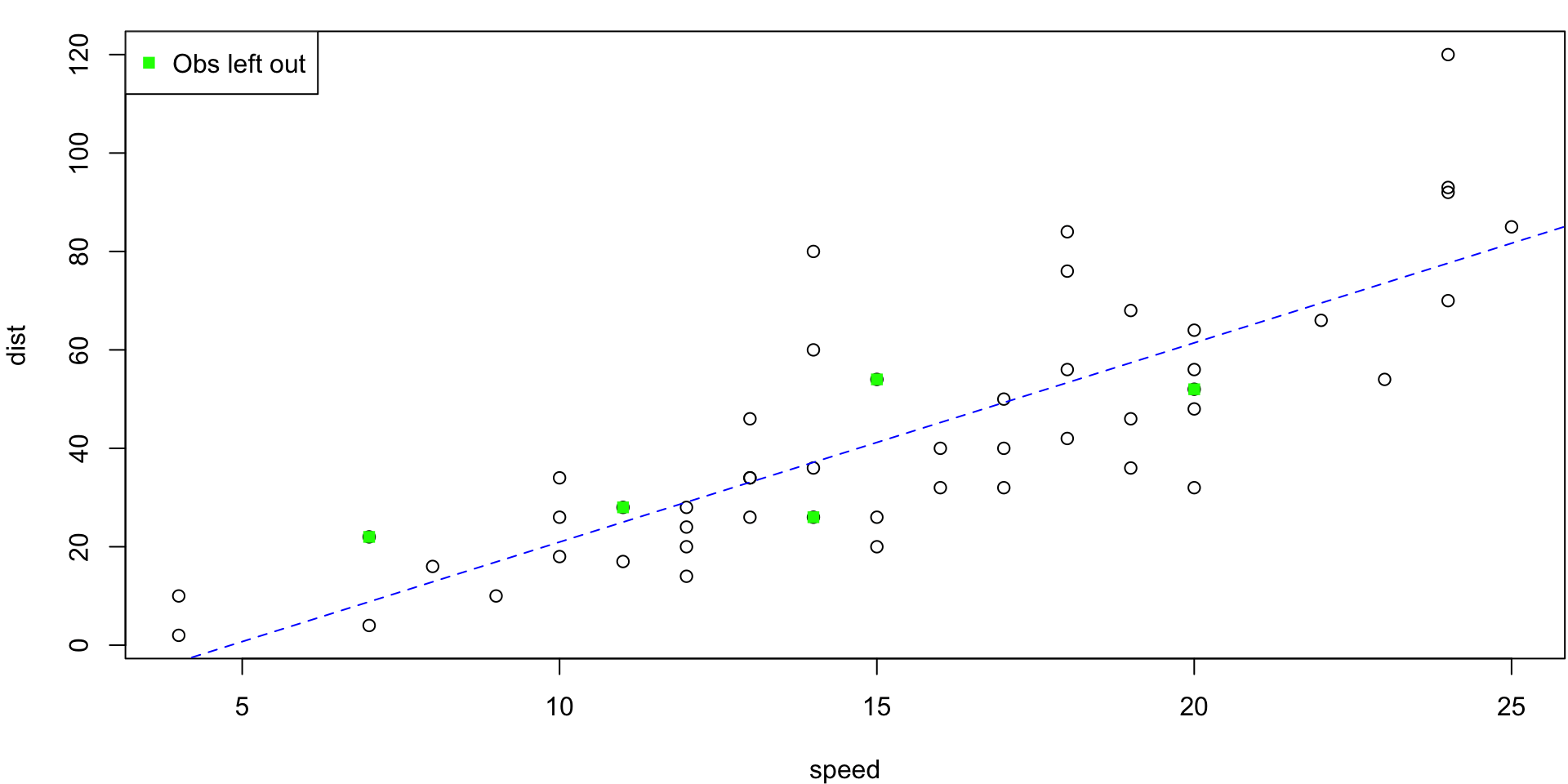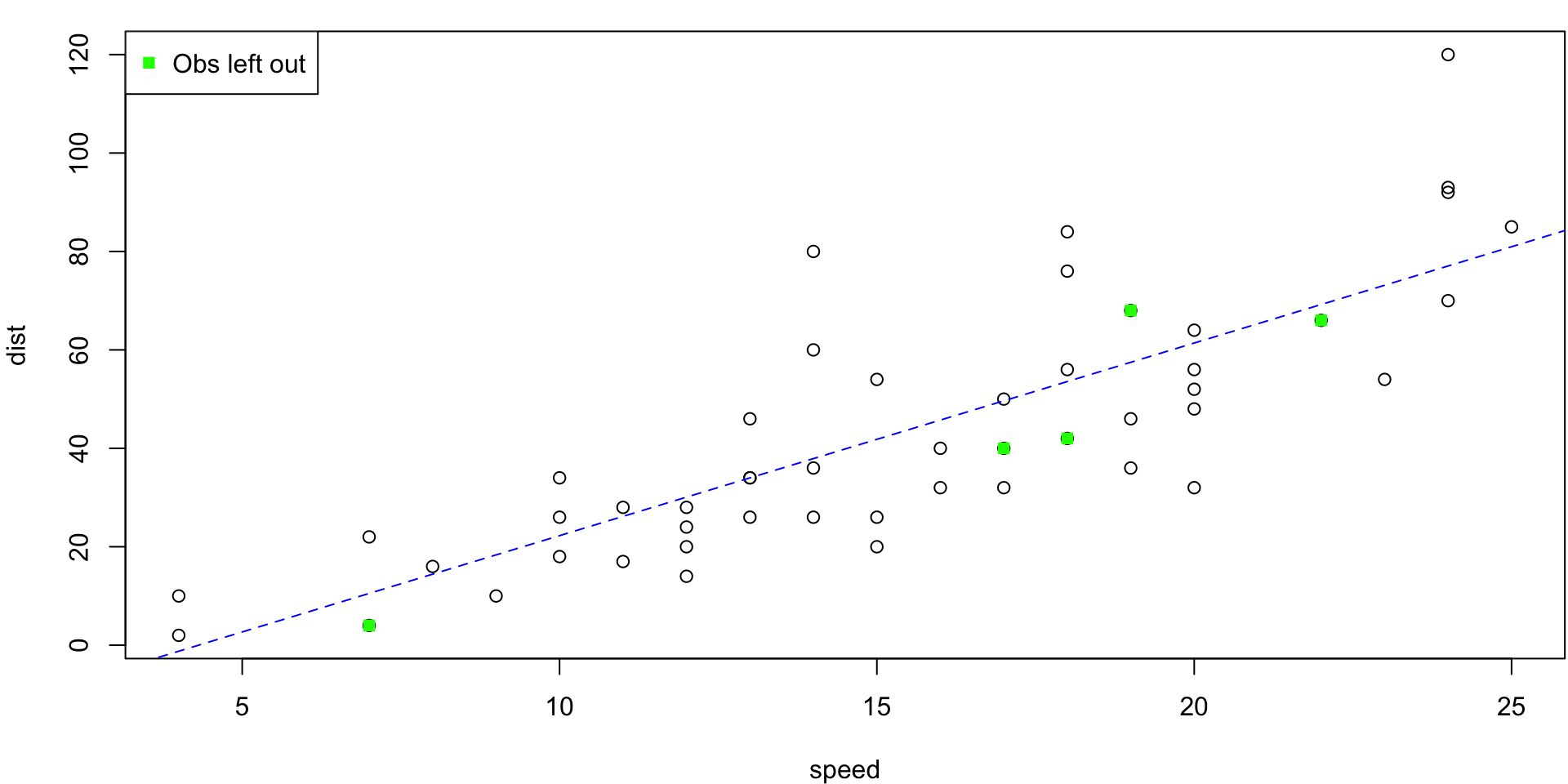
10-fold estimates MSE as \(CV_{(10)} = 243.7798735\)

Summary
| \(k\) | \(CV_{(k)}\) |
|---|---|
| 5 | 254.04 |
| 10 | 243.78 |
| \(n\) (LOOCV) | 246.41 |

Wine example
We now consider the
winedata set from the pgmm package27 measurements on 178 samples of red wine.
Samples originate from the same region of Italy (Piedmont), but are of different varietals (Barolo, Barbera, Grignolino).
We can do KNN classification, using LOOCV1 to choose
k, the number of neighbours…
Best k
In this case the minimum is at k = 8

Wine Example
We can provide a cross-validated classification table,
Predicted
|
|||
|---|---|---|---|
| 1 | 2 | 3 | |
| Barolo | 52 | 3 | 2 |
| Grignolino | 2 | 59 | 11 |
| Barbera | 5 | 9 | 35 |
or LOOCV Error …
\[ \begin{align} CV_{(n)} &= \frac{1}{n} \sum_i^n \text{Err}_i\\ & = \frac{1}{n} \sum_i^n I(y_i \neq \hat y_i)\\ & = \frac{32}{178} = 0.18 \end{align} \]
… or cross-validated versions of any of our other classification performance indices exist!
Standard Errors
More generally: \(CV_K = \sum_{k=1}^K \frac{n_k}{n} \text{Err}_k\)
In addition to computing \(CV_K\) estimate of the error rate, we can also estimate standard deviation of \(CV_K\).
The estimated standard deviation of \(CV_K\) is given by:
Final Remark
- Now we have \(n\) different fits of the model, which model should we report?
None of them
While CV is used to help select a model, the actual reported model would be the one run with all the data.
That is, we would need to run
knn()withk = 8on the full data set with no observations removed.