ANOVA
STAT 205: Introduction to Mathematical Statistics
University of British Columbia Okanagan
Introduction
At this point, we know how to perform inference for the difference of population means from two samples.
To determine if the means from more than two populations are equal we use Analysis of Variance or ANOVA.
ANOVA1 can be considered an extension the (pooled) two-sample \(t\) procedure to more than two groups. However, it involves a test statistic falling a new distribution …

Outline
In this lecture we will be covering
Review of Pooled t-test
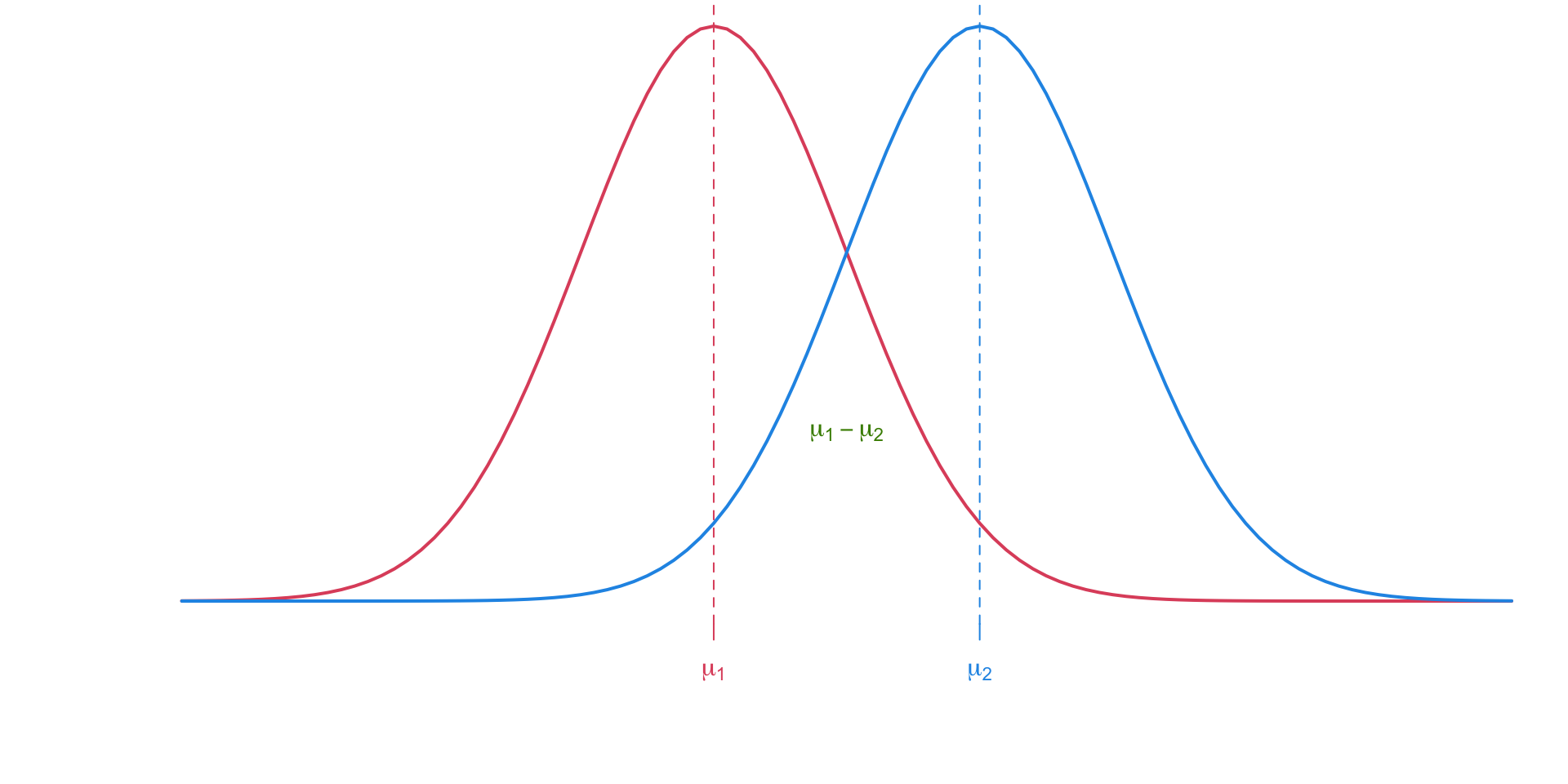
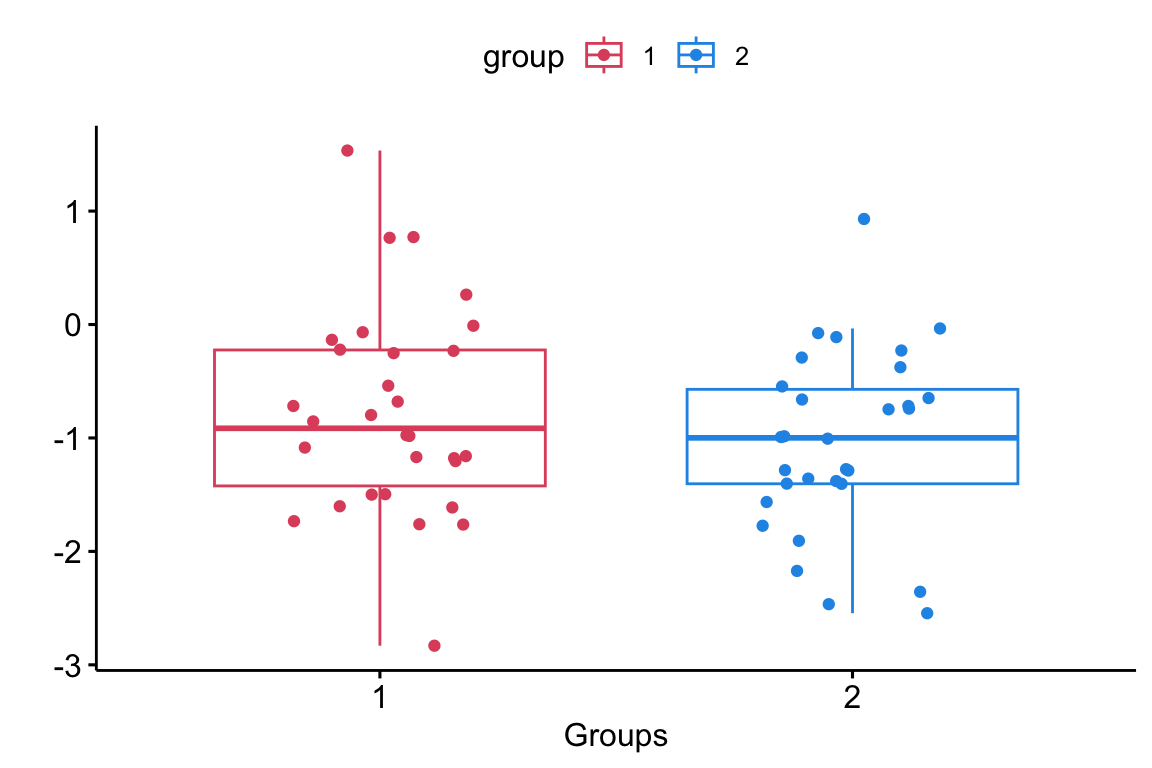
The pooled \(t\)-test assumes equal variance amoung two independent populations. Inference is done on the difference between the two population means \(\mu_1 - \mu_2\).
Pooled Null Hypothesis
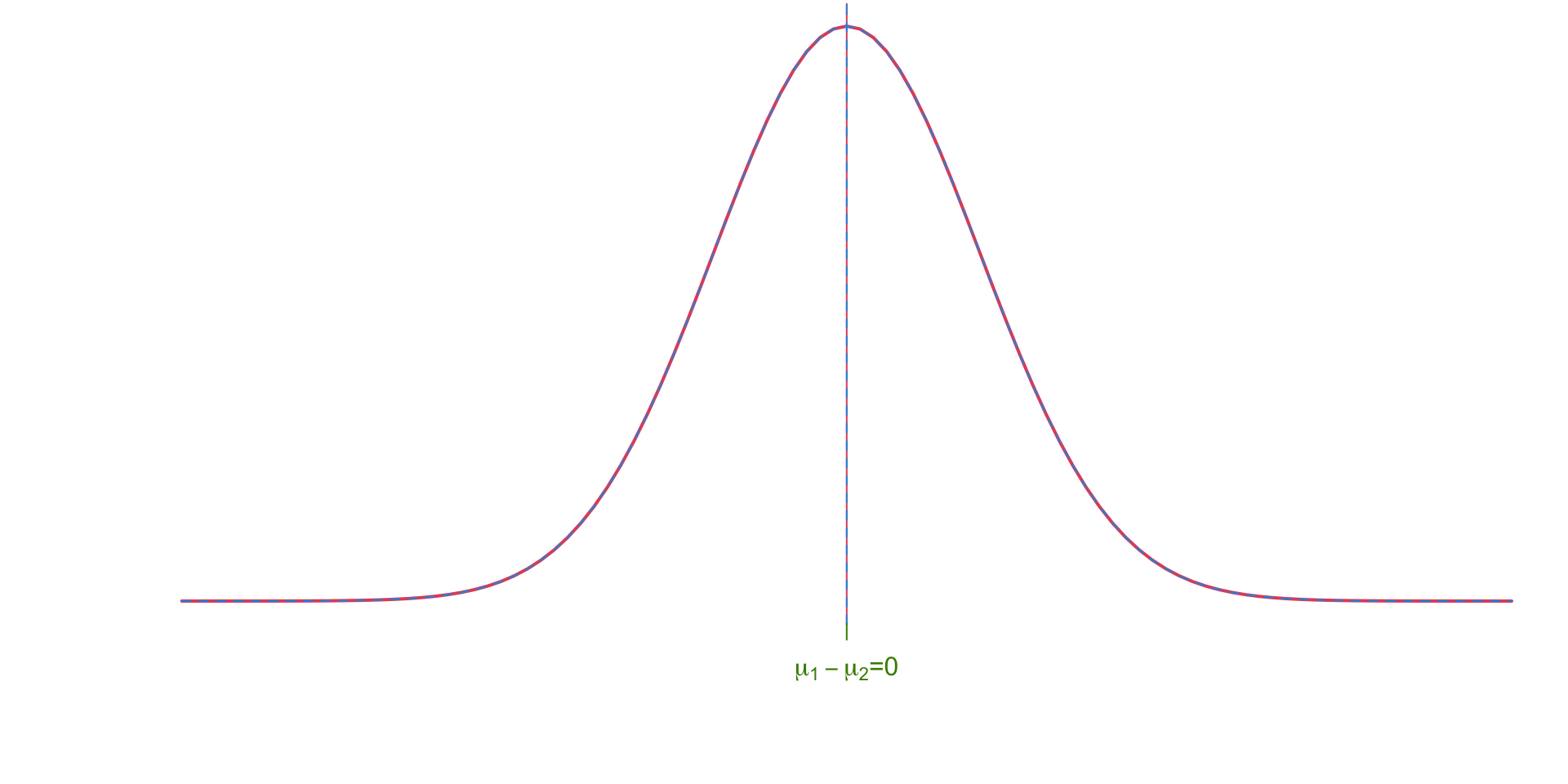
The null hypothesis assumes that there is no difference between the means of the two groups (i.e. \(\mu_1 - \mu_2 = 0)\).
Sample data
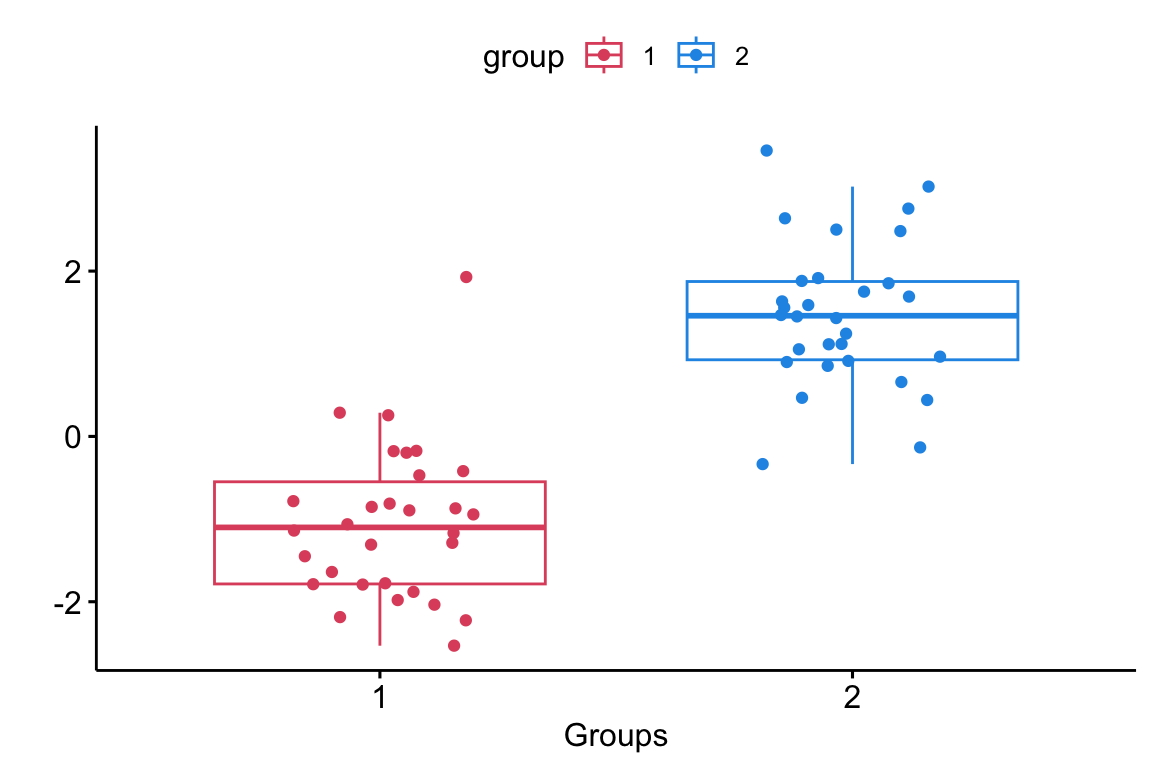
Comparing more than 2 groups

Now we seek to determine whether there are any statistically significant differences between the means of three or more independent groups.
ANOVA null hypothesis
As with the pooled \(t\)-test, our null hypothesis will assume that all group means are equal. The ANOVA test will also require the equal population variance assumption.
Assumptions
In words and symbols, the assumptions of this so-call one-way ANOVA is similar to the pooled \(t\)-test:
- The observations are independent simple random samples from the populations
- The populations are normally distributed
- The population variances are equal \[\sigma_1^2 = \sigma_2^2 = \dots = \sigma_k^2\]
Preamble
The name ANOVA: Analysis of Variance is a bit misleading.
Unlike it’s name suggests, ANOVA is a statistical methods used to investigate differences among the means.
The test involves separating observed variance data into different components and assess whether the observed differences in the sample means are larger than would be expected due to random variation alone.
Example
Let’s examine the rationale behind ANOVA using the following example …
Tar content
Exercise 1 We want to see whether the tar contents (in milligrams) for three different brands of cigarettes are different. Two different labs took samples, Lab Precise and Lab Sloppy.
Hypotheses
-
The null hypothesis in ANOVA asserts that there is no significant difference among the means of the groups being compared, i.e.:
\[H_0: \mu_1 = \mu_2 = \mu_3 = \ldots = \mu_k\]
where \(\mu_1, \mu_2, \mu_3, \ldots, \mu_k\) are the population means of the \(k\) groups being compared.
-
The alternative hypothesis in ANOVA suggests that at least one of the group means is significantly different from the others, i.e.:
\[H_A: \text{ At least one } \mu_i \text{ is different from the others, }\]
where \(i = 1, 2, 3, ..., k\).
Data
Code
library(tidyverse)
# Create the data
lab_precise <- data.frame(
Sample = 1:6,
Brand_A = c(10.21, 10.25, 10.24, 9.80, 9.77, 9.73),
Brand_B = c(11.32, 11.20, 11.40, 10.50, 10.68, 10.90),
Brand_C = c(11.60, 11.90, 11.80, 12.30, 12.20, 12.20)
)
lab_sloppy <- data.frame(
Sample = 1:6,
Brand_A = c(9.03, 10.26, 11.60, 11.40, 8.01, 9.70),
Brand_B = c(9.56, 13.40, 10.68, 11.32, 10.68, 10.36),
Brand_C = c(10.45, 9.64, 9.59, 13.40, 14.50, 14.42)
)
# Calculate the mean of Brand_C in both datasets
mean_brand_c_precise <- mean(lab_precise$Brand_C)
mean_brand_c_sloppy <- mean(lab_sloppy$Brand_C)
# Calculate the adjustment needed for Brand_C in both datasets
adjustment_precise <- 11.5 - mean_brand_c_precise
adjustment_sloppy <- 11.5 - mean_brand_c_sloppy
# Adjust Brand_C values in lab_precise
lab_precise$Brand_C <- lab_precise$Brand_C + adjustment_precise
# Adjust Brand_C values in lab_sloppy
lab_sloppy$Brand_C <- lab_sloppy$Brand_C + adjustment_sloppy
precise_stats <- lab_precise %>%
select(-Sample) %>%
summarise_all(mean)
sloppy_stats <- lab_sloppy %>%
select(-Sample) %>%
summarise_all(mean)
overall_mean = mean(unlist(lab_precise[,-1]))Normality Check for Lab Sloppy

Figure 1: To check our normality assumption we could do a qq-norm plot. Note that we will have to do this for each group in Lab Sloppy
Normality Check for Lab Precise

Figure 2: To check our normality assumption we could do a qq-norm plot. Note that we will have to do this for each group in Lab Precise
Normality Check
We could have checked normality using Shapiro Wilks.
Warning
For large samples, this test often gives small p-values even for minor deviations from normality. For small samples, this test is not very powerful.
✏️ Therefore, use visual aids as your main diagnostics, and use the Shapiro-Wilk test to supplement.
Homogeneity
The equal variance assumption can be checked using:
Graphical Methods ✅ If the spreads (IQRs) of side-by-side boxplots look similar across groups
Rule of Thumb ✅ Largest SD \(\leq\) 2 \(\times\) Smallest SD
-
Statistical Tests1
- Levene’s Test (most common)
- Bartlett’s Test (only use if confident the data are normal)
Hypotheses
While the hypotheses can by written in symbols using: \[\begin{align} H_0:& \mu_1 = \mu_2 = \dots = \mu_k & H_A:& \mu_i \neq \mu_j \text{ for some $i\neq j$} \end{align}\] they are often expressed in plain language as:
- \(H_0\): All group means are equal
- \(H_A\): not all means are equal
- OR \(H_A\): at least one mean is different (more common)
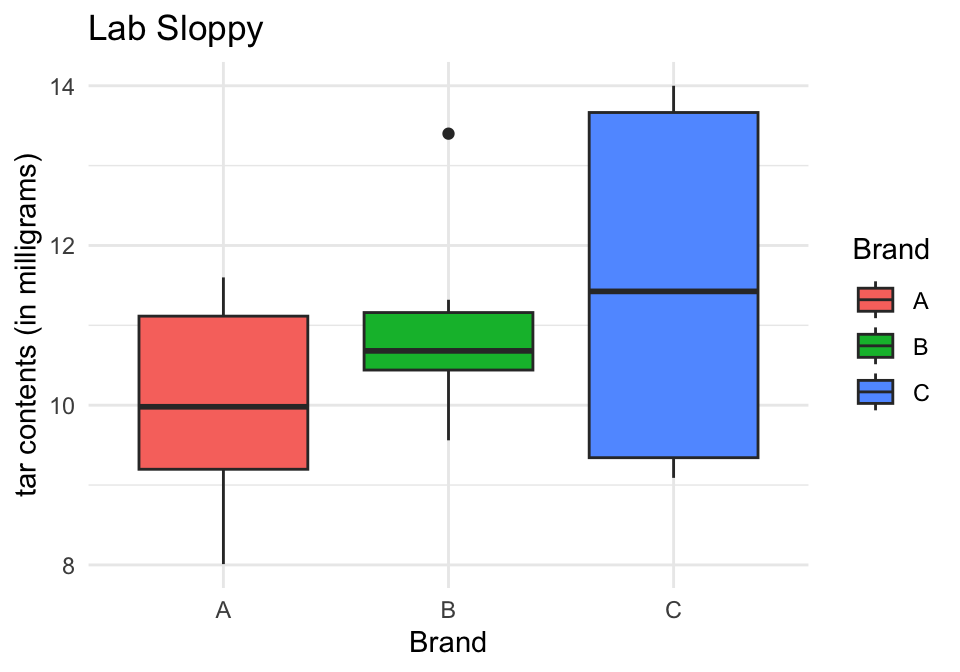
Boxplots Sloppy Labs
Would you guess that the difference in means for the “Sloppy” labs is significantly different?
iClicker
- Yes
- No
- I have no idea
Boxplots Precise Labs
How about for the Precise Lab? Would you guess the difference in means is statistically significant?
iClicker
- Yes
- No
- I have no idea

Boxplots

This sample data is consistent with \(H_0: \mu_A = \mu_B = \mu_C\);
it produces a non-significant \(p\)-value = 0.345451

This sample data provides sufficient evidence against \(H_0: \mu_A = \mu_B = \mu_C\) in favour of the alternative; it produces a significant \(p\)-value = 1.301632e-06
Comments
The difference between these two samples is apparent in the variability within groups, i.e. the boxplots from Lab Sloppy are much wider than Lab Precise.
Since the between-sample-variation from Lab Sloppy is large compared to the within-sample-variation for data from Lab Precise, we will be more inclined to conclude that the three population means are different using the data from Lab Precise.
In order to do an ANOVA test, we will be comparing the between-sample-variation to the within-sample-variation.
Notation
| Group | Data | Mean |
|---|---|---|
| 1 | \(y_{11}, y_{12}, \dots, y_{1n_1}\) | \(\bar y_{1\cdot}\) |
| 2 | \(y_{21}, y_{22}, \dots, y_{2n_2}\) | \(\bar y_{2\cdot}\) |
| \(\vdots\) | \(\vdots\) | \(\vdots\) |
| \(k\) | \(y_{k1}, y_{k2}, \dots, y_{kn_k}\) | \(\bar y_{3\cdot}\) |
- \(k\) total number of groups
- \(y_{ij}\) the \(j\)th observation from the \(i\)th group
- \(n_i\) the sample size from the population
- \(n = \sum_{i=1}^k\) the total sample size
- \(\bar y_{i\cdot}\) the sample mean from the \(i\)th population, i.e. \(y_{i\cdot} = \frac{1}{n_i} \sum_{j=1}^{n_i} y_{ij}\), for \(i = 1, \dots, k\)
- \(\bar y_{\cdot\cdot}\) the overall mean, i.e. \(\bar y_{\cdot\cdot} = \frac{1}{n}\sum_{i = 1}^k \sum_{j=1}^{n_i} y_{ij}\)
- overall sample variance \(s^2 = \frac{1}{n - 1}\sum_{i=1}^{k}\sum_{j=1}^{n_i} (y_{ij} - \bar y_{\cdot\cdot})^2\)
- group sample variance \(s_i^2 = \frac{1}{n_i - 1}\sum_{j=1}^{n_i} (y_{ij} - \bar y_{i\cdot})^2\)
Lab Sloppy
Left: Dot plot showing the tar levels of the sample from Lab Sloppy; the overall mean \(y_{\cdot\cdot} = 10.83\) is depicted as a horizontal gray line. Right: a plot of the tar levels for each brand of cigarettes, grouped and coloured by brand. The group means \(y_{1\cdot}\), \(y_{2\cdot}\), and \(y_{3\cdot}\), are indicated using the pink, green, and blue horizontal line, respectively.

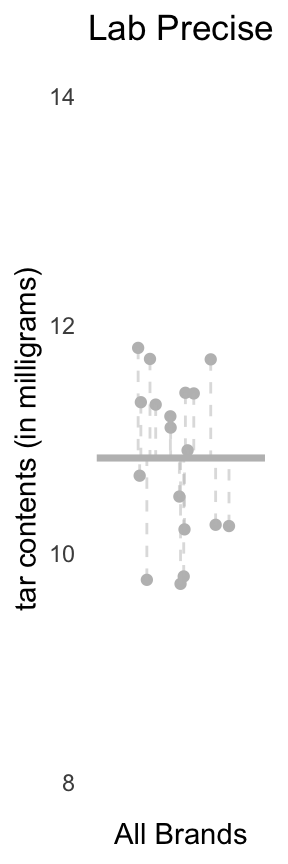
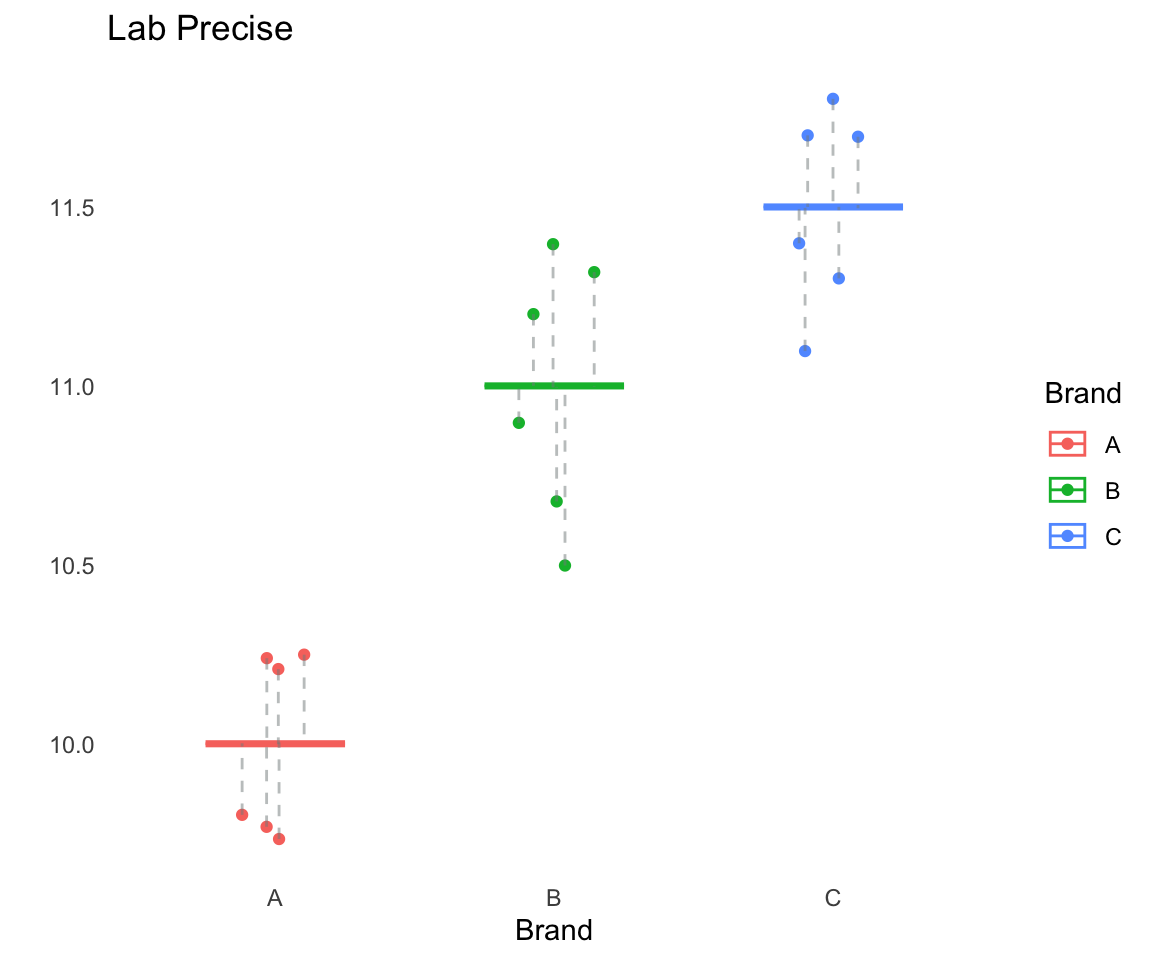
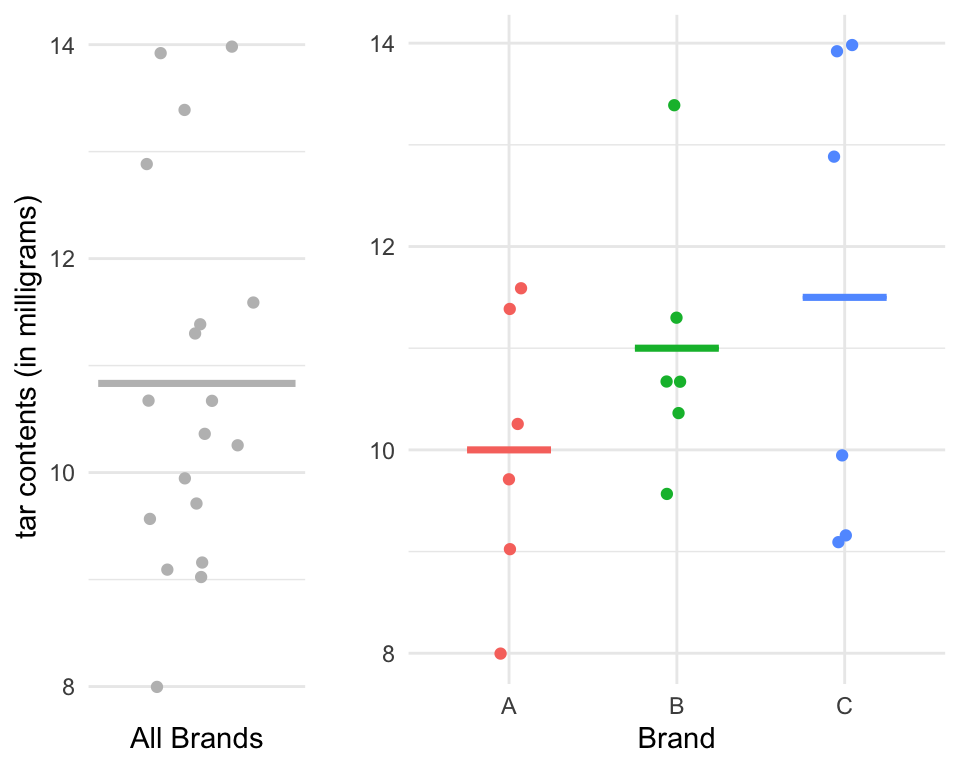
Lab Precise
Left: Dot plot showing the tar levels of the sample from Lab Precise; the overall mean \(y_{\cdot\cdot} = 10.83\) is depicted as a horizontal gray line. Right: a plot of the tar levels for each brand of cigarettes, grouped and coloured by brand. The group means \(y_{1\cdot}\), \(y_{2\cdot}\), and \(y_{3\cdot}\), are indicated using the pink, green, and blue horizontal line, respectively.

Total Sum of Squares

The total sum of squares1 is given by: \[\begin{equation} TSS = \sum_{i,j} (y_{ij} - y_{\cdot \cdot})^2 \end{equation}\]
For Lab Sloppy: \[TSS = 52.9756\]
For Lab Precise: \[TSS = 8.3748\]
Decomposition of TSS
It can be shown that the \(TSS\) can be decomposed into:
- SSW: the within sum of squares, also known as the Sum of Squares for Error and the
- SSB: the between sum of squares, also known as the Sum of Squares for Treatment
\[\begin{equation} TSS = SSW + SSB \end{equation}\]
Definition of SSW and SSB
The SSW is a measure of variation within groups. The formula combines the variability within each sample separately \[ SSW = \sum_{i = 1}^ k \sum_{j = 1}^{n_i} (Y_{ij} - \bar Y_{i\cdot})^2 = \sum_{i,j} (Y_{ij} - \bar Y_{i\cdot})^2 \] The SSB is a measure of the variation between groups. The formula assesses variation in the means from the different samples: \[ SSB = \sum_{i = 1}^{k} \sum_{j = 1}^{n_i} (\bar Y_{i\cdot} - \bar Y_{\cdot\cdot})^2 = \sum_{i = 1}^{k} n_i (\bar Y_{i\cdot} - \bar Y_{\cdot\cdot})^2 \]
Lab Sloppy: Within Sum of Squares
\[\begin{align} SSW &= \sum_{i,j} (y_{ij}- \bar y_{i\cdot})^2\\ &= 45.9756 \end{align}\]
where
- \(\bar y_{1\cdot} = 10\)
- \(\bar y_{2\cdot} = 11\)
- \(\bar y_{3\cdot} = 11.5\)

Lab Precise: Within Sum of Squares
\[\begin{align} SSW &= \sum_{i,j} (y_{ij}- \bar y_{i\cdot})^2\\ &= 1.3748 \end{align}\]
where
- \(\bar y_{1\cdot} = 10\)
- \(\bar y_{2\cdot} = 11\)
- \(\bar y_{3\cdot} = 11.5\)
Lab Sloppy: Between Sum of Squares
\[\begin{align} SSB &= \sum_{i=1}^k n_i(\bar y_{i\cdot}- \bar y_{\cdot\cdot})^2\\ &= 6(10 - 10.83)^2 \\ &+6(11- 10.83)^2 \\ &+6(11.5- 10.83)^2 \\ &= 7 \end{align}\]
\[\begin{align} TSS&= 52.9756\\ &= SSB + SSW\\ &= 7 + 45.9756\\ \end{align}\]

Lab Precise: Between Sum of Squares
\[\begin{align} SSB &= \sum_{i=1}^k n_i(\bar y_{i\cdot}- \bar y_{\cdot\cdot})^2\\ &= 6(10 - 10.83)^2 \\ &+6(11- 10.83)^2 \\ &+6(11.5- 10.83)^2 \\ &= 7 \end{align}\]
\[\begin{align} TSS&= 8.3748\\ &= SSB + SSW\\ &= 7 + 1.3748\\ \end{align}\]

Comments
- SSB captures the variation between the sample means that can be “explained” by differences in \(\mu_i\)s; when the population means differ significantly SSB will be large1
- SSW represents the “unexplained” variation, as it captures the variation within each sample that remains even if the null hypothesis is true.
- If the explained variation, SSB, is significantly larger than the unexplained variation SSW, we should reject \(H_0\).
Total Variance
Recall that we want to examine the between group variation and the within group variation. You may notice that the TSS given by: \[\begin{equation} TSS = \sum_{i,j} (y_{ij} - \bar y_{\cdot \cdot})^2 \end{equation}\] is the numerator of the sample variance of all the sample data, i.e. \[\begin{equation} \text{Overall Sample Variance} = \frac{TSS}{df_t} \end{equation}\] where \(df_t = n - 1\) is the total Degrees of Freedom. When we divide the total SS by it’s df we get the Mean Squares Total (MST).
Degrees of freedom
- \(SSB\) and \(SSW\) are on difference scales so we can not (yet) directly compare them.
- To address this, we calculate analogous Mean Squares metrics, which are obtained by dividing the sum of squares by its corresponding degrees of freedom, \(df\)1
- The resulting \(MSB\) and \(MSW\) are comparable, allowing us to assess the relative contribution of between-group and within-group variability to the total variability in the data.
Formula Summary
Sum of Squares
The Between group Sum of Squares: \[\begin{equation} SSB = \sum_{i=1}^k n_i(\bar y_{i\cdot}- \bar y_{\cdot\cdot})^2 \end{equation}\] The Within Group Sum of Squares: \[\begin{equation} SSW = \sum_{i = 1}^ k \sum_{j = 1}^{n_i} (Y_{ij} - \bar Y_{i\cdot})^2 := \sum_{i,j} (Y_{ij} - \bar Y_{i\cdot})^2 \end{equation}\] Total Sum of Squares (TSS): \[\begin{align} TSS &= \sum_{i,j} (y_{ij} - y_{\cdot \cdot})^2 = SSB + SSW \end{align}\]
Mean Squares
The mean square for between groups \[\begin{equation} MSB = \dfrac{SSB}{k - 1} = \dfrac{SSB}{df_B} \end{equation}\] The mean square for error is \[\begin{equation} MSW = \dfrac{SSW}{n -k} = \dfrac{SSW}{df_W} \end{equation}\] The mean square total is \[\begin{equation} MST = \dfrac{TSS}{n - 1} = \dfrac{TSS}{df_T} \end{equation}\]
Test Statistic
\[\begin{align} F &= \dfrac{\text{between group variance}}{\text{within group variance}}= \dfrac{{MSB}}{MSW} \end{align}\]
- This test statistic follows a so-called F-distribution.
- Under \(H_0\), this ratio should be close to 11
- On the contrary, large values of large F-values indicate differences between groups.
F-distribution

An \(F\)-distribution has two parameters:
- \(\nu_1\) “numerator” df
- \(\nu_2\) “denominator” df
We denote the r.v. \(F_{\nu_1, \nu_2}\)
For one-way ANOVA:
- \(\nu_1 = df_B\) = Number of groups \(= k - 1\)
- \(\nu_2 = df_W\) = Total observations - Number of groups = \(n - k\)
Example: Test Statistic
Back to Exercise 1, our test statistic (i.e. our \(F\)-ratio = \({MSB}/{MSW}\)) follows an \(F_{2, 15}\):

Large F-score
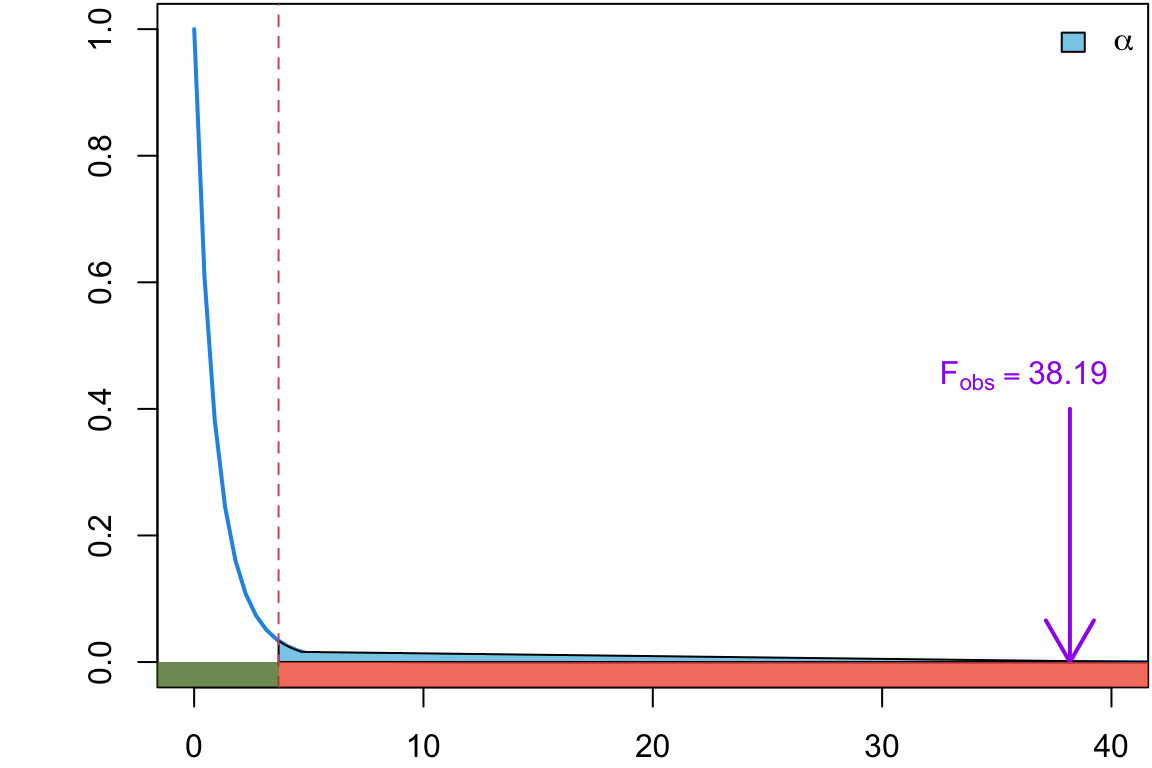
We mentioned that we will reject the null when \(F\) is too big. How big it too big?

Critical Value
How big it too big? \(F_{crit}\) = 3.68

Number Summary
SSB =7, SSW =45.9756,
MSB = 3.5, MSW =3.065 and \[F_{obs} = 1.14\]

SSB =7, SSW =1.3748,
MSB = 3.5, MSW =0.092 and \[F_{obs} = 38.19\]
Decision Summary

p-values
More commonly we will base our decision on a \(p\)-value.
\[ p\text{-value} = \Pr(F_{df_B, df_W} > F_{obs}) \]
Lab Sloppy
\[ \begin{align} &= \Pr(F_{2, 15} > 1.14)\\ &= 0.345451 \end{align} \]
Lab Precise
\[ \begin{align} &= \Pr(F_{2, 15} > 38.19)\\ &= 1.3016316\times 10^{-6} \end{align} \]
p-value plots


Finding F-crit/p-values
While \(F\)-distribution tables exists, they are usually multiple pages long and cumbersome to read off of.
For this course I will not bother with the \(F\)-tables, and will either provide the \(p\)-value for the ANOVA tests, use examples for which the decision is apparent.
Hence, you have some intuition that a very large \(F\)-ratio (e.g. Lab Precise \(F_{obs}\) = 38.19) would produce a very small \(p\)-value, and values close to one (e.g. Lab Sloppy \(F_{obs}\) = 1.14) would yield a large \(p\)-value.
ANOVA table
ANOVA table
| Source | df | SS | MS | F | \(p\)-value |
|---|---|---|---|---|---|
| Between | \(k - 1\) | SSB | MSB | \(\frac{MSB}{MSW}\) | \(\Pr(F_{df_B, df_W} > F_{obs})\) |
| Within | \(n- k\) | SSW | MSW | ||
| Total | \(n-1\) | TSS |
- \(df_T = df_B + df_W\) \(= (k - 1) + (n-k) = n - 1\)
- \(TSS = SSW + SSB\)
- \(MSB = SSB/df_b\), \(MSW = SSW/df_W\)
Example: ANOVA Table
Returning to our Exercise 1, the ANOVA table for Sloppy labs:
| Source | df | SS | MS | F | \(p\)-value |
|---|---|---|---|---|---|
| Between | 2 | 7 | 3.5 | 1.1419101 | 0.345451 |
| Within | 15 | 45.9756 | 3.06504 | ||
| Total | 17 | 52.9756 |
ANOVA table
| Source | df | SS | MS | F | \(p\)-value |
|---|---|---|---|---|---|
| Between | \(k - 1\) | SSB | \(\dfrac{SSB}{k-1}\) | \(\dfrac{MSB}{MSW}\) | \(\Pr(F_{df_B, df_W} > F_{obs})\) |
| Within | \(n- k\) | SSW | \(\dfrac{SSW}{n-k}\) | ||
| Total | \(n-1\) | TSS |
Data format
Very commonly, your data will appear in “wide” format where each column represents a different group (Brand) and the rows represent samples.
From wide to long
In these situations, you would have transform your data to “long” format such that each observation occupies a single row, with the variable(s) represented in columns. l
For our example, each sampled cigarette would appear in the rows and we would have a single column corresponding to Brand (this is a factor with levels:
A,B, orC)We can achieve this in various ways, e.g. using
reshape(),melt()orpivot_longer()functions to name a few.
long table
Note: lab_sloppy and lab_precise defined on Data slide
library(tidyverse)
# Reshape the data from wide to long format
sloppy_long <- pivot_longer(lab_sloppy, cols = starts_with("Brand"),
names_to = "Brand", values_to = "Value")
# Modify the "Brand" column to only show the letters 'A', 'B', and 'C'
# i.e. change Brand_A, Brand_B, Brand_C to A, B, C
sloppy_long <- sloppy_long %>%
mutate(Brand = substr(Brand, nchar(Brand), nchar(Brand)))
# same for Lab Precise:
precise_long <- pivot_longer(lab_precise, cols = starts_with("Brand"),
names_to = "Brand", values_to = "Value") %>%
mutate(Brand = substr(Brand, nchar(Brand), nchar(Brand)))ANOVA in R
You can fit an analysis of variance model in R using aov()
the first argument is the formula which has the following form:
-
response_variable= variable whose variation you are interested in explaining or predicting based on the grouping variable. -
group_variable= the categorical variable that defines the groups or categories in your data. Each level of the grouping variable represents a different group/population -
~separates the left-hand side (response variable) from the right-hand side (predictors)
Sloppy ANOVA Table
Returning to our Exercise 1, the ANOVA table for Sloppy labs becomes:
| Source | df | SS | MS | F | \(p\)-value |
|---|---|---|---|---|---|
| Between | 2 | 7 | 3.5 | 1.1419101 | 0.345451 |
| Within | 15 | 45.9756 | 3.06504 | ||
| Total | 17 | 52.9756 |
And in R…
Precise ANOVA Table
Returning to our Exercise 1, the ANOVA table for Precise labs becomes:
| Source | df | SS | MS | F | \(p\)-value |
|---|---|---|---|---|---|
| Between | 2 | 7 | 3.5 | 38.1873727 | 1.3016316^{-6} |
| Within | 15 | 1.3748 | 0.0916533 | ||
| Total | 17 | 8.3748 |
And in R…
Decision
As usual, if the \(p\)-value is less than our predefined significance level \(\alpha\), we reject the null hypothesis.
Reject \(H_0 \implies\) conclude that not all the means are equal: that is, at least one mean is different from the other means.
Notice there is no indication on which mean or means are statistically different (is one or multiple brands were better/worse than another?).
In Exercise 1, a follow up question might be which brands differ in average tar content.
Multiple Comparison
To answer this follow-up question we conduct an analysis of all possible pairwise means.
This involves a method called multiple comparisons. 1
-
For Exercise 1, with three brands of cigarettes, A, B, and C, we would compare the three possible pairwise comparisons:
Brand A to Brand B \(H_0: \mu_A = \mu_B\)
Brand A to Brand C \(H_0: \mu_A = \mu_C\)
Brand B to Brand C \(H_0: \mu_B = \mu_C\)
Summary
ANOVA is a formal hypothesis test for multiple means that compares the within variation and between variation.
The one-way ANOVA showed statistically significant results for Lab Precise but not for Lab Sloppy.
Lab Precise: the within variation was small compared to the between variation thus resulted in a large F-statistic (38.1873727) and thus a small p-value (1.301632e-06).
Lab Sloppy: this ratio was small (1.1419101), resulting in a large p-value (0.345451).
References
Material from this lecture has been adapted from “Statistics Online: Courses, Resources, and Materials. (n.d.). Retrieved from https://online.stat.psu.edu/stat500/”
Comment about assumptions
Non-extreme departures from the normality assumes are not that serious since the sampling distribution of the test statistic is fairly robust, especially as sample size increases sample sizes for all groups are equal.
If you conduct an ANOVA test, you should always try to keep the same sample sizes for each group; this is known as balanced study design.
A general rule of thumb for equal variances is to compare the smallest and largest sample standard deviations. This is much like the rule of thumb for equal variances for the test for independent means. If the ratio of these two sample standard deviations falls within 0.5 to 2, then it may be that the assumption is not violated.