# Install caret if you haven't already
install.packages("caret")Cross-validation and the Bootstrap
Cross-validation and the Bootstrap
Introduction
In this lab, you will learn how to implement cross-validation and bootstrapping techniques in R to assess model performance and improve model robustness. These methods are essential for evaluating models beyond a single training set, helping to prevent overfitting and giving you more reliable estimates of model accuracy.
Goal: Learn how to perform k-fold cross-validation and leave-one-out cross-validation (LOOCV) to evaluate the performance of regression models. We will look at “by-hand” code and some functions from packages
R packages
Although these things are easy enough to code up by hand, we’ll be using some helpful pacakges to streamline the resampling and cross-validation approach.
carat package
The caret package (short for Classification And REgression Training) contains miscelleanous functions for training and plotting classification and regression models. For the detailed documentation visit: https://topepo.github.io/caret/. The current release version can be found on CRAN/caret and the project is hosted on github. Before we begin make sure you’ve installed the package using:
Recall, you should run this line directly in your console and you need only do it once. Each new session we will have to call and load the library using:
library(caret)Loading required package: ggplot2Loading required package: latticetrainControl function
The trainControl() function in the caret package determines how the model training will be conducted. In this function we will need to specify the resampling method as either "boot" (for bootstraping), "cv" (for \(k\)-fold cross validation), "LOOCV" (for leave-one-out cross validation), or "none" (only fits one model to the entire training set). Note: there are more options that you can read about in the documentation; see ?trainControl
train function
The train() function
# General Syntax:
train(formula, data, method, trControl = NULL, tuneGrid = NULL, tuneLength = NULL, ...)The main arguments are:
formula: The model formula (e.g.,dist ~ speedfor regression).data: The dataset to be used for training.method: The machine learning algorithm you want to use. For example,lmfor linear regression,rffor random forests,knnfor k-nearest neighbors, etc…1trControl: A list specifying the type of resampling or validation to use. This is created using thetrainControl()function and can define cross-validation, bootstrapping, or other methods.tuneGrid: A custom grid of hyperparameters that you can provide for tuning the model. This is particularly useful when you want to manually specify the values to try.tuneLength: An integer that defines how many different values of the hyperparameters should be tested. If notuneGridis provided,tuneLengthis used to automatically generate a range of values for tuning....: Additional arguments specific to the model.
createFolds function
The createFolds() function in the caret package is used to split your data into subsets or “folds” for the purpose of k-fold cross-validation. It creates indices that can be used to divide your dataset into training and testing subsets repeatedly, depending on how many folds you specify.
# Syntax
createFolds(y, k = 10, list = TRUE, returnTrain = FALSE)The main arguments are:
y: A vector of data (usually the response variable or class labels) that is used to ensure stratified sampling. The function tries to create folds such that the proportion of different class labels or response values are roughly the same across all folds.k: The number of folds (or subsets) you want to divide the data into. The default isk = 10, which creates 10 folds.list: A logical value (default isTRUE) that specifies whether the output should be returned as a list. Iflist = TRUE, the function returns a list of indices for each fold. Iflist = FALSE, it returns a vector indicating the fold assignment for each observation.returnTrain: A logical value (default isFALSE). IfTRUE, the function returns the training set indices for each fold (i.e., all indices except the ones in that fold). IfFALSE, it returns the test set indices for each fold.
Output:
When
list = TRUE: The function returns a list where each element is a vector of indices corresponding to one fold. These indices can be used to subset your data for training or validation purposes.When
list = FALSE: The function returns a vector where each element corresponds to a fold assignment for each observation
boot package
The boot packages contains functions and datasets for bootstrapping from the book “Bootstrap Methods and Their Application” by A. C. Davison and D. V. Hinkley (1997, CUP), originally written by Angelo Canty for S. The current release version can be found on CRAN/boot. The documentation can be found here: https://cran.r-project.org/web/packages/boot/boot.pdf. It also contains handy functions and datasets for bootstrapping from the book Bootstrap Methods and Their Applicationby A. C. Davison and D. V. Hinkley (1997, CUP)
# Install caret if you haven't already
install.packages("boot")Recall, you should run this line directly in your console and you need only do it once. Each session, you’ll need to load the library before using any of its functions:
library(boot)
Attaching package: 'boot'The following object is masked from 'package:lattice':
melanomaThe cv.glm() function from the boot package in R is used to perform cross-validation for generalized linear models (GLMs), including ordinary linear models (lm). This function is versatile and can be used for different types of cross-validation, such as k-fold cross-validation and Leave-One-Out Cross-Validation (LOOCV), to estimate the predictive error of a model.
# General Syntax
cv.glm(data, glmfit, cost = function(y, yhat) mean((y - yhat)^2), K = n)The main arguments are:
data: The data frame that contains the variables used in the model.glmfit: The fitted model object created using theglm()function orlm()(since linear models are a special case of GLMs). This is the model on which cross-validation will be performed.cost: This is a function that defines the cost function to be minimized. By default, the cost is the Mean Squared Error (MSE), but you can specify other loss functions depending on your needs.K: The number of groups into which the data should be split to estimate the cross-validation prediction error2. The default is to setKequal to the number of observations indatawhich gives the usual leave-one-out cross-validation.
The returned value is a list with the following components.
call |
The original call to cv.glm. |
K |
The value of K used for the K-fold cross validation. |
delta |
A vector of length two. The first component is the cross-validation estimate of prediction error3. |
seed |
The value of .Random.seed when cv.glm was called. |
Cars data
For a quick demo on how to do CV we will use the cars dataset from class. For simplicity, I will assume the entire cars data set makes up our training set so \(n\) in this case is 50.
nrow(cars)[1] 50cars data sets
The caret package also has a cars data set. We will be working with the cars data set from the datasets package (which comes preinstalled in R). If the cars dataset has been overwritten after loading the caret package, you can easily reload the original cars dataset from the datasets package using the following code:
data(cars, package = "datasets") Linear Regression
LOOCV
Leave-one-out cross validation (LOOCV) is a special case of \(k\)-fold CV when \(k =n\), the number of observations in our training set.
Manual coding
It’s relatively easy to set-up leave-one-out cross-validation yourself for any analysis. Here we can do it for the simple linear model…
# load the data set: specify the package to
# avoid confusion with the cars data from caret
data(cars, package = "datasets")
attach(cars)
lm_loocv <- list()
mseloocv <- NA
for(i in 1:nrow(cars)){
cvspeed <- speed[-i]
cvdist <- dist[-i]
lm_loocv[[i]] <- lm(cvdist ~ cvspeed)
mseloocv[i] <- (predict(lm_loocv[[i]], newdata=data.frame(cvspeed=speed[i])) - dist[i])^2
}
# Calculate the CV MSE test
loocv_mse_slr = mean(mseloocv)
loocv_mse_slr[1] 246.4054\[MSE_{CV_{(n)}} = \frac{1}{n} \sum_{i=1}^n MSE_i = 246.4\]
Note: I am using slight different notation than lecture to distinguish between some other cross-validated metrics herein.
Using the caret package
Alternatively, this could be done using the caret package; see boot package section for general usage and syntax:
# Load necessary libraries
library(caret)
# Set up the trainControl function for LOOCV
control_loocv <- trainControl(method = "LOOCV")
# Run lm for LOOCV
lm_loocv <- train(dist ~ speed, data = cars, method = "lm", trControl = control_loocv)
# View results
lm_loocvLinear Regression
50 samples
1 predictor
No pre-processing
Resampling: Leave-One-Out Cross-Validation
Summary of sample sizes: 49, 49, 49, 49, 49, 49, ...
Resampling results:
RMSE Rsquared MAE
15.69731 0.6217139 12.05918
Tuning parameter 'intercept' was held constant at a value of TRUENotice that by default the regression metric don’t include the MSE. To calculate the CV MSE, we can simply square the CV RMSE:
# Extract RMSE from the model's results (this is the cross-validation RMSE)
rmse_loocv <- lm_loocv$results["RMSE"]
# Calculate MSE by squaring the RMSE
mse_loocv <- rmse_loocv^2
mse_loocv <- unname(mse_loocv) # remove the RMSE label
mse_loocv
1 246.4054\[ MSE_{CV_{(n)}} = (RMSE_{CV_{(n)}})^2 = 246.405416^2 = 246.405416 \]
which, unsurprising, agrees with the one we calculated by hand. Recall: LOOCV is deterministic (i.e. not random) so we should get the same answer each time.
Using boot
You can also use the boot package to directly calculate cross-validation error (MSE) for an lm model. The cv.glm() function performs cross-validation and returns the MSE.
# Fit a linear model
lm_model <- glm(dist ~ speed, data = cars)
# the above is equivalent to
# lm_model <- lm(dist ~ speed, data = cars)
# default K = n (so we can just leave K unspecified)
loocv_results <- cv.glm(cars, lm_model)
# Extract the cross-validated MSE
loocv_mse <- loocv_results$delta[1] # First value is the cross-validated MSE
# Print cross-validated MSE
loocv_mse[1] 246.4054Again, notice, that this estimate is consistent across each method (as it should be).
k-fold Cross-validation
For \(k\)-fold cross validation, the dataset is divided into \(k\) parts/folds. At iteration 1, the first fold is used as the testing set and the remaining \(k-1\) folds is used as the training set. The process is repeated \(k\) times with each of the \(k\) folds serving as the test set once. The out-of-sample performance, i.e. the performance metrics (e.g., accuracy, mean squared error, ROC-AUC) computed on each of the \(k\) test sets, are averaged to produced the CV estimate.
semi-manual
While it would be relatively easy to code up \(k\)-fold CV validation ourselves, we can leverage the work of several packages to speed things up. For instance, in order to create \(k\) folds we can use the createFolds() function from the caret package. For instance, if we wanted to do 5-fold cross validation:
# choose the number of folds
k = 5# Create K folds
set.seed(123)
folds <- createFolds(cars$speed, # a vector of outcomes
k = k) # the number of folds desiredThe above will create k folds of indices from y = cars$speed. Each fold will contain indices for the data points that belong in the fold. We can view this using:
str(folds)List of 5
$ Fold1: int [1:9] 2 7 8 19 25 31 35 39 50
$ Fold2: int [1:9] 1 3 9 18 26 34 36 41 45
$ Fold3: int [1:12] 6 13 14 16 20 24 28 33 37 40 ...
$ Fold4: int [1:11] 11 12 15 22 23 29 32 38 43 46 ...
$ Fold5: int [1:9] 4 5 10 17 21 27 30 42 44Here, folds$Fold1 will return the indices of the data points that are in the first fold, folds$Fold2 will give the second fold, and so on…
Next, we will train the model on each of the \(k\) different training sets, where each set leaves out a single fold for testing. Note that folds[i] returns a list (even if it contains only one element). To convert this list into a simple vector of indices, we use unlist().
mse_values <- numeric(k) # a vector to store MSEi's
for (i in 1:k) {
train_indices <- unlist(folds[-i]) # Training indices
test_indices <- unlist(folds[i]) # Test indices
# Fit the model on the training set
model <- lm(dist ~ speed, data = cars[train_indices,])
# or we could have coded this up as:
# model <- lm(dist ~ speed, data = cars subset = train_indices)
# Make predictions on the test set
predictions <- predict(model, newdata = cars[test_indices,])
# Calculate the Mean Squared Error (MSE)
mse_values[i] <- mean((cars$dist[test_indices] - predictions)^2)
}
# Cross-Validated MSE estimate:
(cv_mse_k5 <- mean(mse_values))[1] 240.3434For completion of the table below I’ve rerun the code with \(k\)=10 (hidden to save space and avoid repetition)
Code
k = 10
# Create K folds
set.seed(10)
folds <- createFolds(cars$speed, k = k)
mse_values <- numeric(k)
for (i in 1:k) {
train_indices <- unlist(folds[-i]) # Training indices
test_indices <- unlist(folds[i]) # Test indices
model <- lm(dist ~ speed, data = cars[train_indices,])
predictions <- predict(model, newdata = cars[test_indices,])
mse_values[i] <- mean((cars$dist[test_indices] - predictions)^2)
}
cv_mse_k10 <- mean(mse_values)k-fold using the carat package
To perform k-fold cross-validation using the caret package, you can use the train() function, which simplifies the process of cross-validation for a wide variety of models. The trainControl() function allows you to specify the resampling method, such as k-fold cross-validation.
You can specify how many folds you want to use by setting the method to "cv" (for cross-validation) and setting number to the number of folds you want. We’ll use 5 and 10 for demonstration purposes (normally you can choose just one).
# Define the cross-validation method
control_5fold <- trainControl(method = "cv", number = 5)
control_10fold <- trainControl(method = "cv", number = 10)Next we train the model with cross-validation using the train(). The example below uses a linear regression model (lm), but you can replace lm with other model types.
# Train the model using 5-fold cross-validation
set.seed(9894)
model5 <- train(dist ~ speed, data = cars, method = "lm", trControl = control_5fold)
# Train the model using 10-fold cross-validation
set.seed(1463)
model10 <- train(dist ~ speed, data = cars, method = "lm", trControl = control_10fold)Notice how I use different seeds. Using different seeds in code when training models, such as in cross-validation, can help introduce variety into the random processes involved, such as partitioning data into folds, shuffling, or selecting subsets. If you use the same seed each time in your code, it will result in the same random sequence being generated for each operation that involves randomness.
The results will include the cross-validated performance of the model, such as the RMSE (Root Mean Squared Error) or other relevant metrics depending on the model. As before, we will convert these to CV MSE:
# Extract the cross-validation RMSE:
rmse_cv5 <- model5$results["RMSE"]
rmse_cv10 <- model10$results["RMSE"]
# Calculate MSE by squaring the RMSE
mse_cv5 <- unname(rmse_cv5^2)
mse_cv5
1 239.847mse_cv10 <- unname(rmse_cv10^2)
mse_cv10
1 230.585k-fold using the boot package
The cv.glm() function from the boot package calculates the estimated K-fold cross-validation prediction error for generalized linear models. Recall that these models have a shortcut formula so the model need only be fit once to obtain the CV estimates for MSE. To use the function we must first fit our regression model using glm()4:
The code is similar to that which we did in the LOOCV section above, only now, instead of using K = \(n\) (default), we need to choose a reasonable value for K, see Note 1. For demonstration purposes, we’ll do 5-fold and 10-folds:
# Define the linear model
glm_model <- glm(dist ~ speed, data = cars)
# 5-fold cross-validation
set.seed(123)
cv_results_5 <- cv.glm(cars, glm_model, K = 5)
cv_results_10 <- cv.glm(cars, glm_model, K = 10)Unlike LOOCV, the first value returned by delta is the cross-validated MSE:
cv_mse_5 <- cv_results_5$delta[1]
cv_mse_5[1] 244.1175cv_mse_10 <- cv_results_10$delta[1]
cv_mse_10[1] 242.0437KNN regression
Next we’ll do the same exercise for KNN. We’ll do this three ways: once using knn.reg(), once using caret. Note that the boot package is best suited for performing CV with linear models, so it is not appropriate here. Since KNN requires the tuning parameter of \(k\) nearest neighbours (which will get confusing since we also use k to denote the number of folds!) we will also need to specify which values of \(k\) to try. To be consistent with the previous section, lets try from 1 to 49. To avoid confusion between the two \(k\)’s well use k_folds and k_nn where possible.
LOOCV KNN
knn.reg
knn.reg() from the FNN package is another example of a function that has built-in (LOO) cross validation; see ?knn.reg. As described in the help file, if test is not supplied, Leave one out cross-validation is performed. Here we can make use of that to choose the number of nearest neighbours for KNN regression.
library(FNN)
kr <- list()
predmse <- NA
# the candidate values to try for our number of nearest neighbours
k_nn_vec = 1:49
for(i in k_nn_vec){
kr[[i]] <- knn.reg(speed, test=NULL, dist, k=i)
predmse[i] <- kr[[i]]$PRESS/nrow(cars)
}
plot(predmse, type="l", lwd=3, col="red")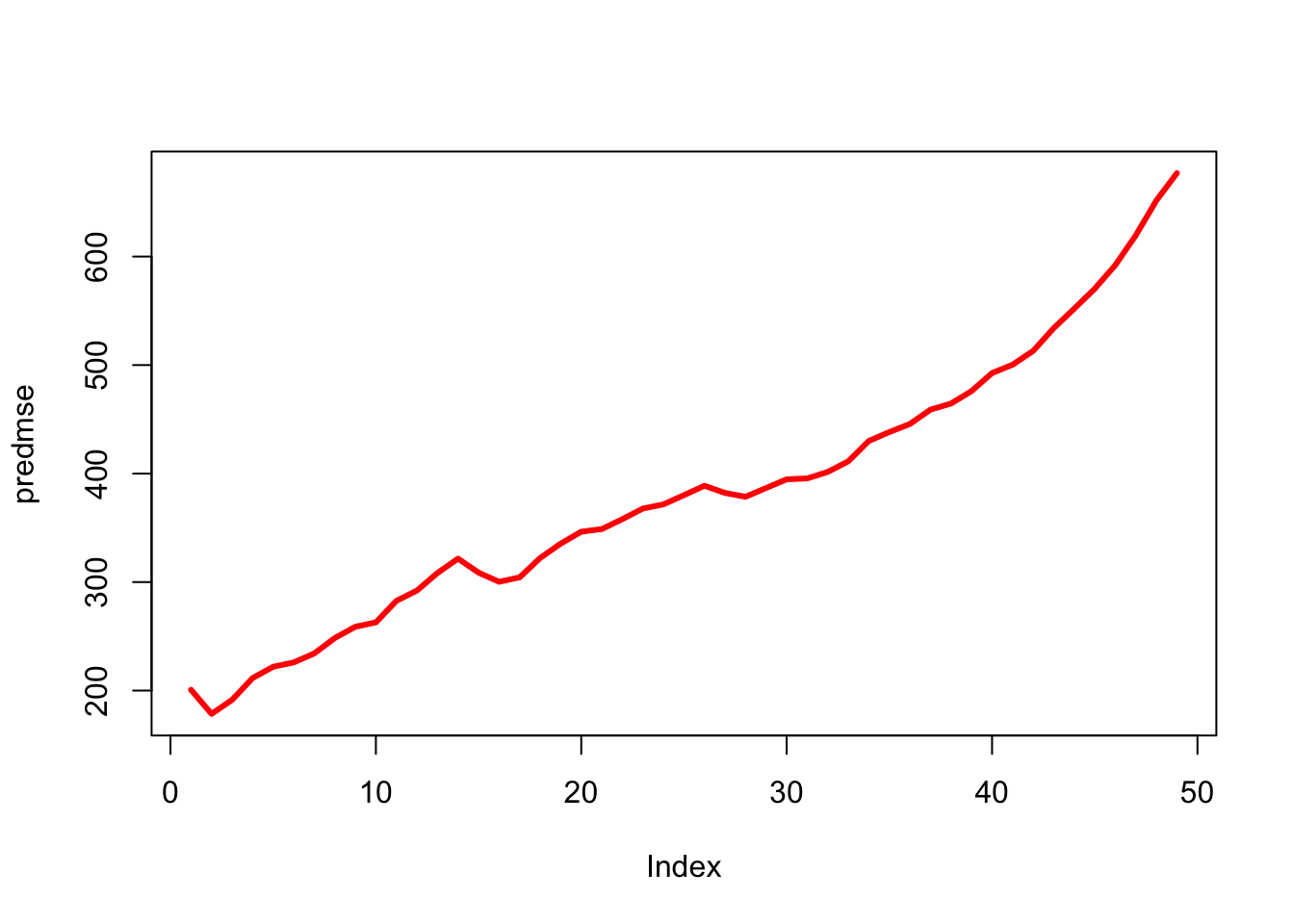
which.min(predmse)[1] 2plot(dist~speed)
seqx <- seq(from=0, to=30, by=0.01)
knnr <- knn.reg(speed, y=dist, test=matrix(seqx, ncol=1), k=2)
lines(seqx, knnr$pred, col="green", lwd=1)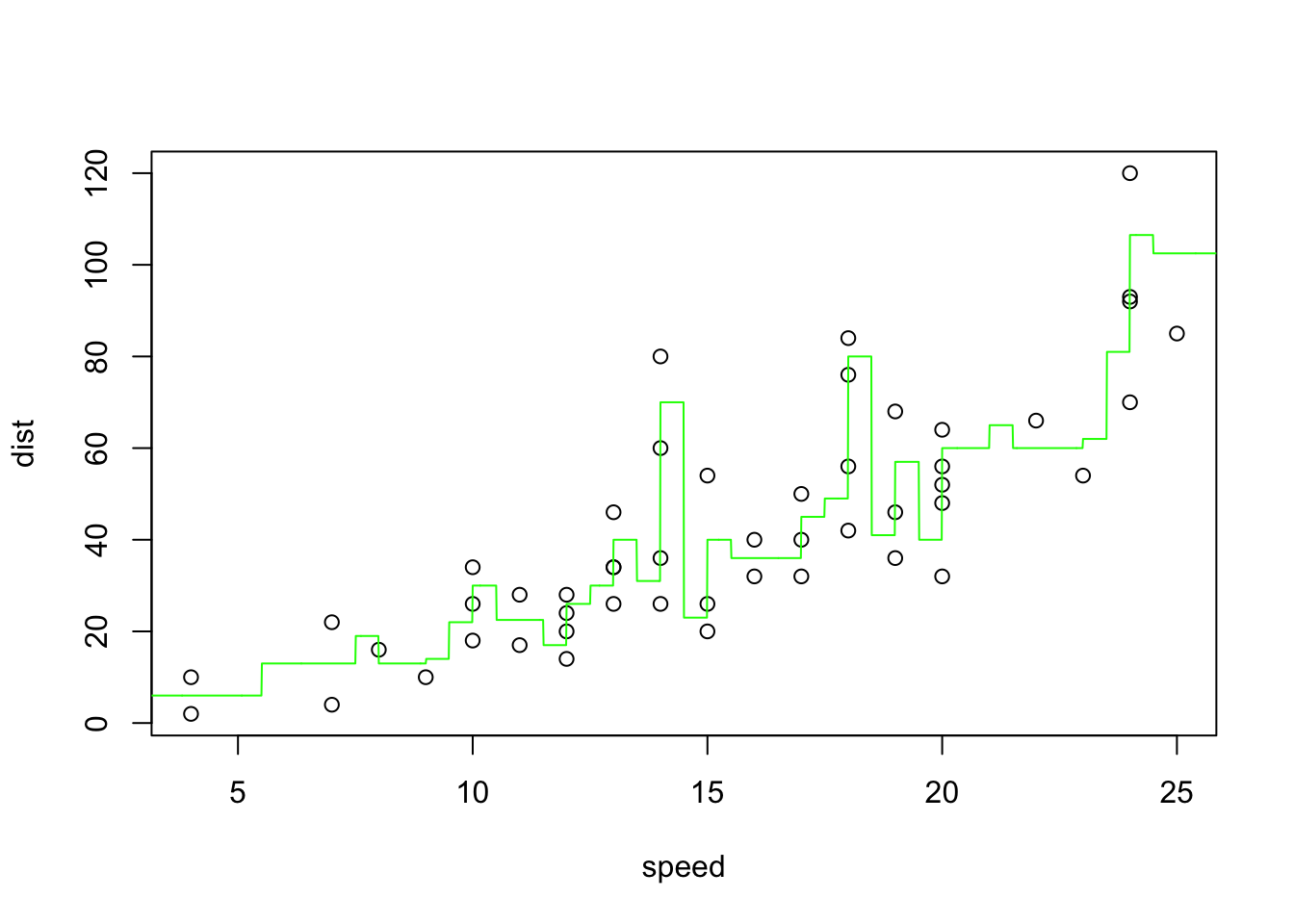
The CV MSE for the winning model is:
(mse_knn_reg = predmse[which.min(predmse)])[1] 178.535caret KNN
The code for this section is similar to the LOOCV in the linear regression section, only now, we need to switch the method from lm to knn .
# Set up training control using LOOCV
train_control <- trainControl(method = "LOOCV")
# Create a grid of k values from 1 to 49
k_grid <- expand.grid(k = 1:49)
# Define the model using k-NN
set.seed(123)
knn_model_loocv <- train(dist ~ speed,
data = cars,
method = "knn",
trControl = train_control,
tuneGrid = k_grid) # Trying all k values from 1 to 49
# View the results (the best model is stroed as)
knn_model_loocvk-Nearest Neighbors
50 samples
1 predictor
No pre-processing
Resampling: Leave-One-Out Cross-Validation
Summary of sample sizes: 49, 49, 49, 49, 49, 49, ...
Resampling results across tuning parameters:
k RMSE Rsquared MAE
1 17.22299 0.57771971 13.84900
2 16.81462 0.59364378 13.03469
3 16.32874 0.62188660 12.74524
4 15.98970 0.60869934 12.27888
5 15.79924 0.61692667 11.96067
6 15.98720 0.60793955 12.26667
7 16.84921 0.58456213 12.75144
8 17.06976 0.56319622 13.03896
9 16.85434 0.58027828 12.82896
10 17.16124 0.56586393 13.27007
11 17.37702 0.55581363 13.17682
12 17.77312 0.54363718 13.30913
13 17.73366 0.55443281 13.30204
14 17.79687 0.55225609 13.39734
15 17.93630 0.55212054 13.56614
16 17.89203 0.55650024 13.48466
17 18.22349 0.52581857 13.78783
18 18.21679 0.52824000 13.73147
19 19.06778 0.49151984 14.51179
20 18.95363 0.49988875 14.44539
21 18.94276 0.50118566 14.42743
22 19.09070 0.49595241 14.68826
23 19.03806 0.51229277 14.55499
24 19.48921 0.48796762 14.75037
25 19.59459 0.48054593 14.80893
26 19.66843 0.48332166 14.97472
27 19.76024 0.48379838 14.98436
28 19.86830 0.48500253 15.09865
29 20.08624 0.47776134 15.51675
30 19.89567 0.49971544 15.38104
31 20.86988 0.46895544 16.13642
32 20.85333 0.47125698 16.08570
33 20.85647 0.48008157 16.15815
34 20.84894 0.48089403 16.13155
35 21.71419 0.47208035 16.78270
36 21.77776 0.46328199 16.93608
37 21.76089 0.46493056 16.91877
38 22.12245 0.44183983 17.31407
39 22.46702 0.41400914 17.49862
40 22.50325 0.40066240 17.64702
41 22.91034 0.38323974 17.91136
42 22.89386 0.38930235 17.91045
43 23.04356 0.39556208 18.20231
44 23.39353 0.36641378 18.51179
45 24.92671 0.17613614 20.20738
46 25.33215 0.04315454 20.53483
47 25.35466 0.03169222 20.55194
48 25.78441 0.36798681 20.82569
49 26.03100 1.00000000 21.11918
RMSE was used to select the optimal model using the smallest value.
The final value used for the model was k = 5.As displayed in the output, the best value of \(k\) as chosen by cross validation is 5. You can get that answer programatically using:
# Extract the best k value
best_k_loocv <- knn_model_loocv$bestTune
cat("Best k (LOOCV):", best_k_loocv$k, "\n")Best k (LOOCV): 5 Note that the final model that is trained using the best \(k\) value is stored in the finalModel component. You can access it like this:
knn_model_loocv$finalModel5-nearest neighbor regression modelHmm… we’re getting difference answers here than in LOOCV KNN. I’m not sure why that is… I will need to investigate this further.
As before, we can get the cross-validated estimate of MSE by squaring the cross-validated estimate for RMSE. This can be achieved using:
# Extract cross-validated RMSE (Root Mean Squared Error)
win_index = which.min(knn_model_loocv$results$RMSE)
rmse_loocv <- knn_model_loocv$results$RMSE[win_index]
# Calculate MSE by squaring the RMSE
mse_loocv <- rmse_loocv^2
mse_loocv[1] 249.6161Finally let’s plot the original data and the fitted KNN model with the number of nearest neighbours as choose by CV:
plot(cars$speed, cars$dist, xlab = "Speed", ylab = "Distance",
main = paste("KNN Regression (k =", best_k_loocv, ")"))
# Make predictions using the best model
seqx <- seq(from=0, to=30, by=0.01)
newdata <- data.frame(speed = seqx)
predictions <- predict(knn_model_loocv, newdata = newdata)
lines(seqx, predictions, col = "red", lwd = 2)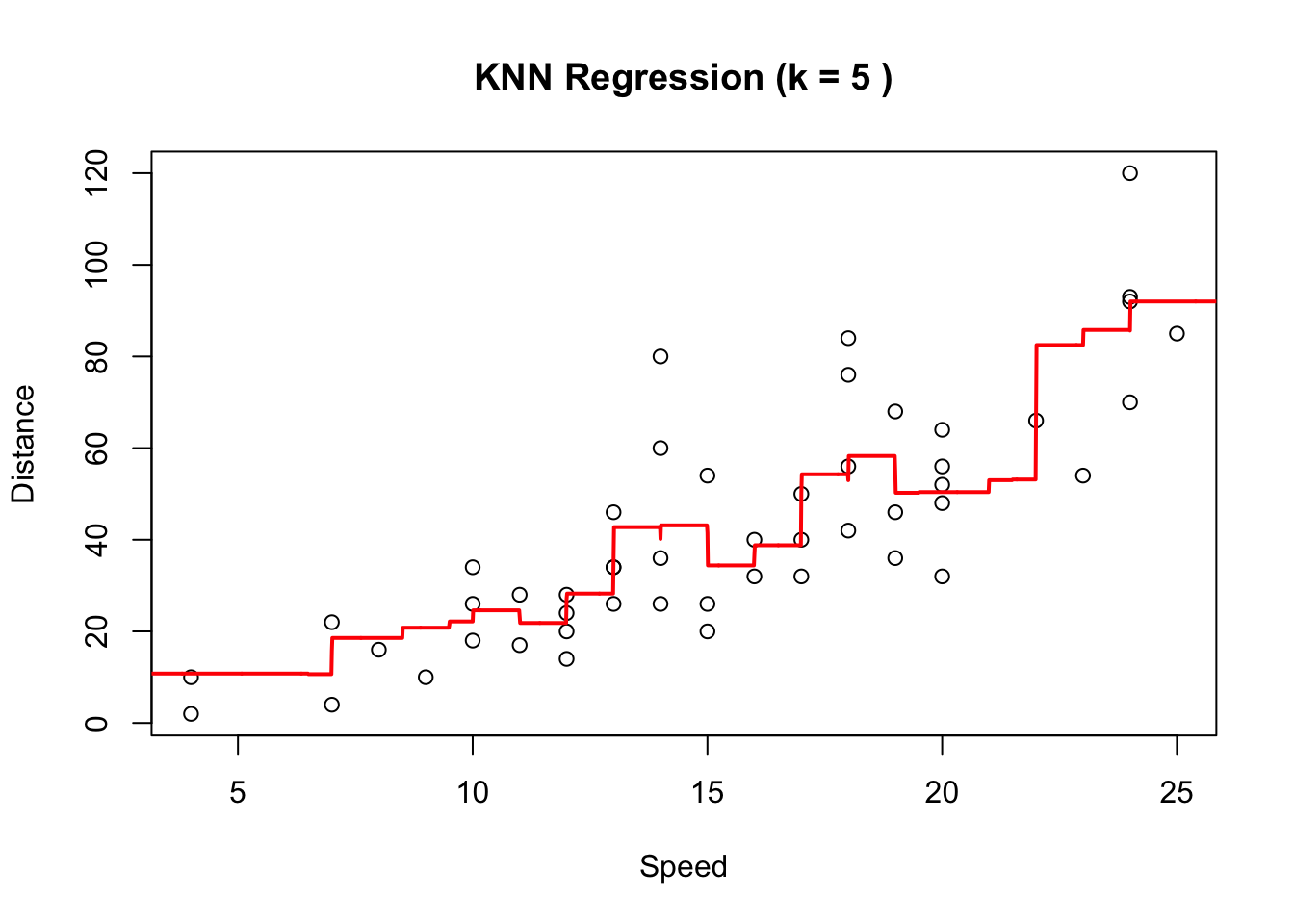
k-fold cross validation
When fitting KNN on the entire dataset, we initially considered the values from 1 through 49. This is no longer feasible for cross-validation because \(k\) cannot exceed the number of points in the training set. When performing cross-validation (CV), each training set is smaller as it excludes the validation fold. Therefore, we’ll adjust the code to test from 1 up to 15.
knn.reg
To perform 5-fold cross-validation using knn.reg from the FNN package for k-Nearest Neighbors regression, you will need to manually set up the cross-validation process, as knn.reg does not have built-in cross-validation functionality like caret does.
As before, we will perform 5-fold and 10-fold cross-validation. If you’ve followed the examples above, the following code should be self-explanatory.
# Set up number of folds
k_folds <- 5
# Create K folds for cross-validation
set.seed(123)
folds <- createFolds(cars$speed, k = k_folds, list = TRUE)
# Initialize a matrix to store the MSE values for each k (1 to 49) across all folds
k_values <- 1:10
mse_matrix5 <- matrix(NA, nrow = k_folds, ncol = length(k_values))
# Loop through each value of k (number of neighbors) to perform cross-validation
for (k in k_values) {
for (i in 1:k_folds) {
# Get training and test indices
train_indices <- unlist(folds[-i])
test_indices <- unlist(folds[i])
# Split the data into training and testing sets
train_data <- cars[train_indices, ]
test_data <- cars[test_indices, ]
# Perform k-NN regression for this value of k
# Convert the 'speed' column to matrix form to ensure knn.reg works correctly
knn_model <- knn.reg(train = as.matrix(train_data$speed),
test = as.matrix(test_data$speed),
y = train_data$dist, k = k)
# Calculate the MSE for this fold and store it
mse_matrix5[i, k] <- mean((test_data$dist - knn_model$pred)^2)
}
}
# Calculate the average MSE across all folds for each value of k
avg_mse_per_k5 <- colMeans(mse_matrix5, na.rm = TRUE)
# Find the optimal k (the one with the lowest MSE)
optimal_k5 <- which.min(avg_mse_per_k5)
cat("Optimal k:", optimal_k5, "\n")Optimal k: 5 cv_mse_5fold = avg_mse_per_k5[optimal_k5]
# Print the MSE for the optimal k
cat("Cross-validated MSE for optimal k:", cv_mse_5fold, "\n")Cross-validated MSE for optimal k: 265.2471 Again using 10-folds.
Code
# Set up number of folds
k_folds <- 10
# Create K folds for cross-validation
set.seed(123)
folds <- createFolds(cars$speed, k = k_folds, list = TRUE)
# Initialize a matrix to store the MSE values for each k (1 to 49) across all folds
mse_matrix10 <- matrix(NA, nrow = k_folds, ncol = length(k_values))
# Loop through each value of k (number of neighbors) to perform cross-validation
for (k in k_values) {
for (i in 1:k_folds) {
# Get training and test indices
train_indices <- unlist(folds[-i])
test_indices <- unlist(folds[i])
# Split the data into training and testing sets
train_data <- cars[train_indices, ]
test_data <- cars[test_indices, ]
# Perform k-NN regression for this value of k
# Convert the 'speed' column to matrix form to ensure knn.reg works correctly
knn_model <- knn.reg(train = as.matrix(train_data$speed),
test = as.matrix(test_data$speed),
y = train_data$dist, k = k)
# Calculate the MSE for this fold and store it
mse_matrix10[i, k] <- mean((test_data$dist - knn_model$pred)^2)
}
}
# Calculate the average MSE across all folds for each value of k
avg_mse_per_k10 <- colMeans(mse_matrix10, na.rm = TRUE)
# Find the optimal k (the one with the lowest MSE)
optimal_k10 <- which.min(avg_mse_per_k10)
cat("Optimal k:", optimal_k10, "\n")Optimal k: 3 Code
cv_mse_10fold = avg_mse_per_k10[optimal_k10]
# Print the MSE for the optimal k
cat("Cross-validated MSE for optimal k:", cv_mse_10fold, "\n")Cross-validated MSE for optimal k: 251.8796 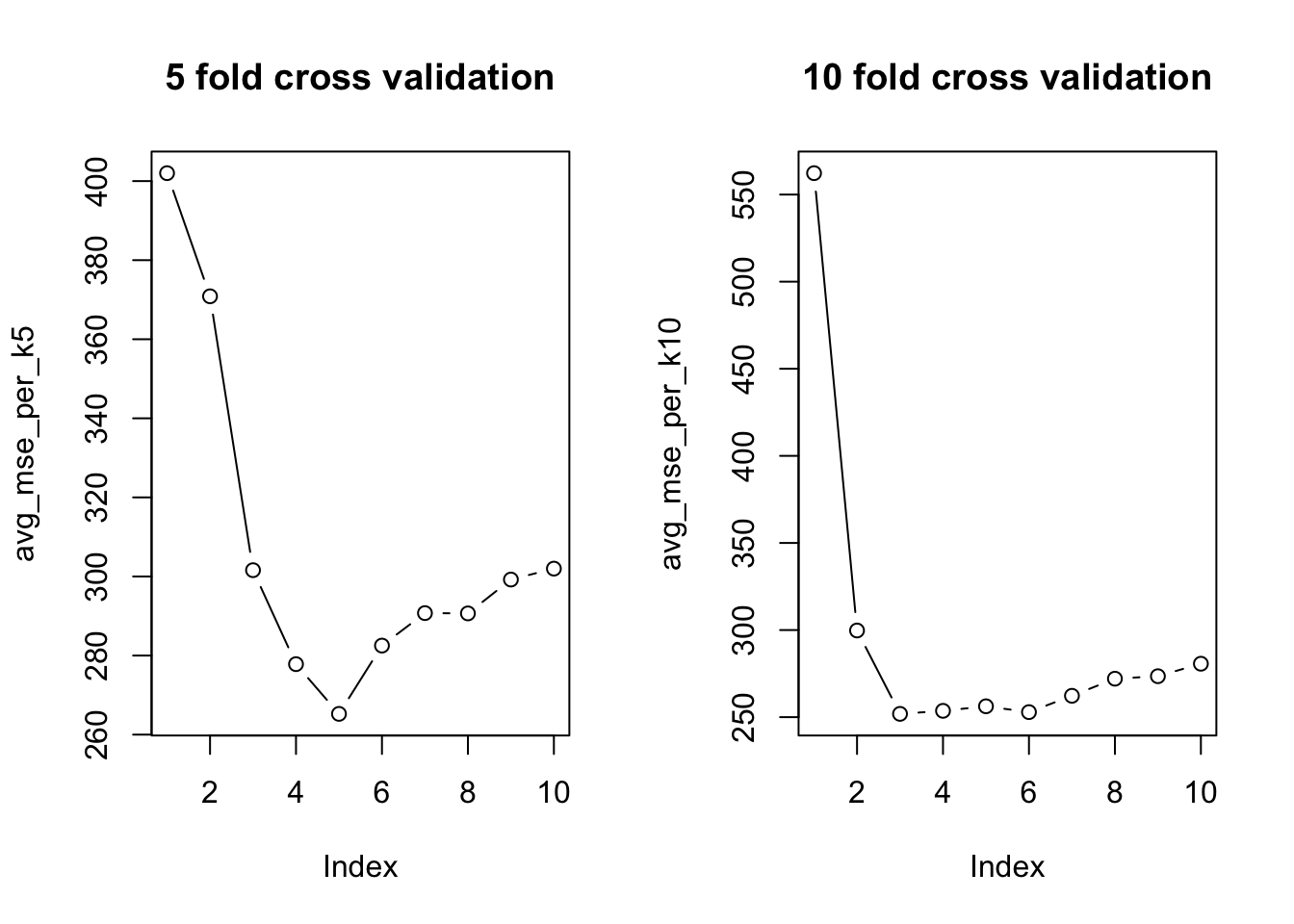
Notice how the optimal choice for \(k\) differs from the 5-fold and 10-fold implementation; the former choose a \(k\) nearest neighbour value of 5 and the latter, \(k\) = 3
caret
To perform K-fold cross-validation and extract the MSE using caret, here’s how you can proceed:
# Set up training control for 5-fold cross-validation
set.seed(5023)
train_control <- trainControl(method = "cv", number = 5)
# Create a grid of k values from 1 to 30
k_grid <- expand.grid(k = 1:10)
# Define the model using k-NN
knn_model_cv <- train(dist ~ speed,
data = cars,
method = "knn",
trControl = train_control,
tuneGrid = k_grid)
# Look at the results
knn_model_cvk-Nearest Neighbors
50 samples
1 predictor
No pre-processing
Resampling: Cross-Validated (5 fold)
Summary of sample sizes: 39, 39, 41, 41, 40
Resampling results across tuning parameters:
k RMSE Rsquared MAE
1 17.82436 0.6099704 14.49827
2 17.05437 0.6403416 13.52805
3 16.40135 0.6662305 12.56247
4 15.48093 0.6812195 11.45096
5 14.96545 0.6773739 11.47145
6 16.11588 0.6330477 12.64884
7 15.84545 0.6504624 12.16476
8 16.11074 0.6355928 12.58649
9 16.57940 0.6005171 12.93264
10 16.55728 0.6134936 12.70252
RMSE was used to select the optimal model using the smallest value.
The final value used for the model was k = 5.# Extract the RMSE for the best model
win_index5 = which.min(knn_model_cv$results$RMSE)
best_rmse5 <- knn_model_cv$results$RMSE[win_index5]
# Convert RMSE to MSE
best_mse5 <- best_rmse5^2
# Print the cross-validated MSE
print(best_mse5)[1] 223.9646Do the same for 10-fold
Code
# Set up training control for 5-fold cross-validation
train_control <- trainControl(method = "cv", number = 5)
# Train KNN regression model using caret with cross-validation
knn_model <- train(dist ~ speed,
data = cars,
method = "knn",
trControl = train_control,
tuneGrid = k_grid)
# Extract the RMSE for the best model
win_ind10 = which.min(knn_model$results$RMSE)
best_rmse10 <- knn_model$results$RMSE[win_ind10]
# Convert RMSE to MSE
best_mse10 <- best_rmse10^2Summary
Below are the summary results from our experiments.
| Method | knn.reg() | caret |
|---|---|---|
| LOOCV | 178.535 (2 ) | 249.62 (5) |
| 5-fold CV | 265.25 (5) | 223.96 (5) |
| 10-fold CV | 251.88 (3) | 231.25 (5) |
Notice how there is some discrepancy on the optimal value for \(k\). See as how 5 appears the most often; I would be inclined to go with that.
Bootstrap Estimation
Manual
Here we will recreate the simulation provided in lecture for the nonparametric bootstrap
set.seed(311532)
x <- runif(30, 0, 1)
y <- 2*x + rnorm(30, sd=0.25)
mod1 <- lm(y~x)
newx <- list()
newy <- list()
modnew <- list()
coefs <- NA
for(i in 1:1000){
#newx[[i]] <- runif(30, 0, 1)
newx[[i]] <- x
newy[[i]] <- 2*newx[[i]] + rnorm(30, sd=0.25)
modnew[[i]] <- lm(newy[[i]]~newx[[i]])
coefs[i] <- modnew[[i]]$coefficients[2]
}
set.seed(35134)
newboots <- list()
bootsmod <- list()
bootcoef <- NA
xy <- cbind(x,y)
for(i in 1:1000){
newboots[[i]] <- xy[sample(1:30, 30, replace=TRUE),]
bootsmod[[i]] <- lm(newboots[[i]][,2]~newboots[[i]][,1])
bootcoef[i] <- bootsmod[[i]]$coefficient[2]
}
summary(mod1)$coefficients Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.00427974 0.08754461 -0.04888639 9.613569e-01
x 2.05167004 0.12940570 15.85455727 1.620243e-15sd(coefs)[1] 0.1325126sd(bootcoef)[1] 0.132347The theoretical standard error is 0.13735, so our hope is that all three values should be close to that. sd(coefs) (aside from the theoretical truth) would be a gold-standard approach for finding that value — but it is of course essentially impossible in practice to repeatedly gather new data. summary(mod1)$coefficients is based on the inferential linear regression assumptions (iid normal error, etc) and only one model fit. sd(bootcoef) is the bootstrap estimator based on resampling (with replacement) from the original sample.
As in class, we can push further by using the observed bootstrap estimators in bootcoef as an empirical distribution. First, we looked at the histogram
hist(bootcoef)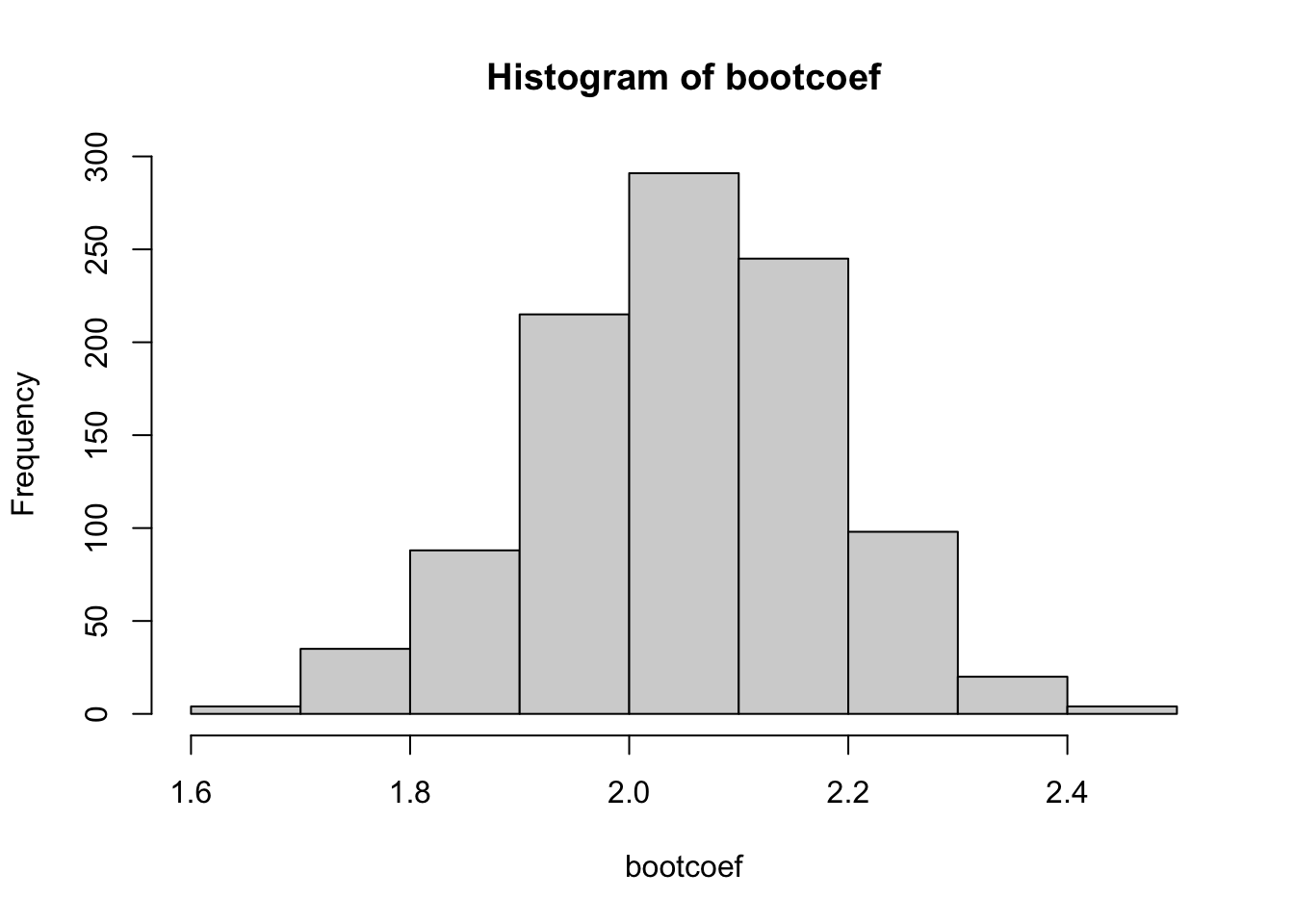
The simplest bootstrap confidence interval, often called Efron intervals (after the bootstrap founder Bradley Efron), simply pull the relevant percentiles of the observed bootstrap estimates.
So for example, for an approx 95% CI we would want the 2.5th and 97.5th percentiles of the observed estimates…
quantile(bootcoef, c(0.025, 0.975)) 2.5% 97.5%
1.766970 2.296553 How about a 90% CI? Well, that would need the 5th and 95th percentiles
quantile(bootcoef, c(0.05, 0.95)) 5% 95%
1.825115 2.261882 Notice that the true theoretical value (2.00) is contained within both of these intervals.
Using exisiting functions
Alternatively we could use existing packages like boot to automate the bootstrapping process. These packages provide functions that simplify the implementation and often support parallel processing for efficiency.
For example we could redo the above example using:
# Load the boot package
library(boot)
# Create a function to calculate the statistic of interest
beta_function <- function(data, indices) {
sample_data <- data[indices,]
fit <- lm(y~x, data = sample_data)
return(coef(fit)["x"])
}
# Number of bootstrap samples
B <- 1000
# Set a seed for reproducibility
set.seed(6663492)
# Perform bootstrapping
dat <- data.frame(x, y)
boot_results <- boot(dat, beta_function, R = B)
# View the results
print(boot_results)
ORDINARY NONPARAMETRIC BOOTSTRAP
Call:
boot(data = dat, statistic = beta_function, R = B)
Bootstrap Statistics :
original bias std. error
t1* 2.05167 0.002535141 0.1267117Here beta_function() fits the linear regression model and pulls out the estimate for slope. More generally we will need to create a function that calculates our statistic (i.e. our function of our data, say, \(T = t(X)\)) out of resampled data. It should have at least two arguments: a data argument and an indices vector containing indices of elements from data that were picked to create a bootstrap sample.
boot_results have a few noteworthy outputs. $t contains the values our statistic \(T(Z^{*j})\) where \(Z^{*j}\) is the bootstrap sample and \(j = 1, 2, \dots, B\):
head(boot_results$t) [,1]
[1,] 2.093083
[2,] 2.070167
[3,] 1.914885
[4,] 1.917636
[5,] 1.936294
[6,] 2.078371hist(boot_results$t)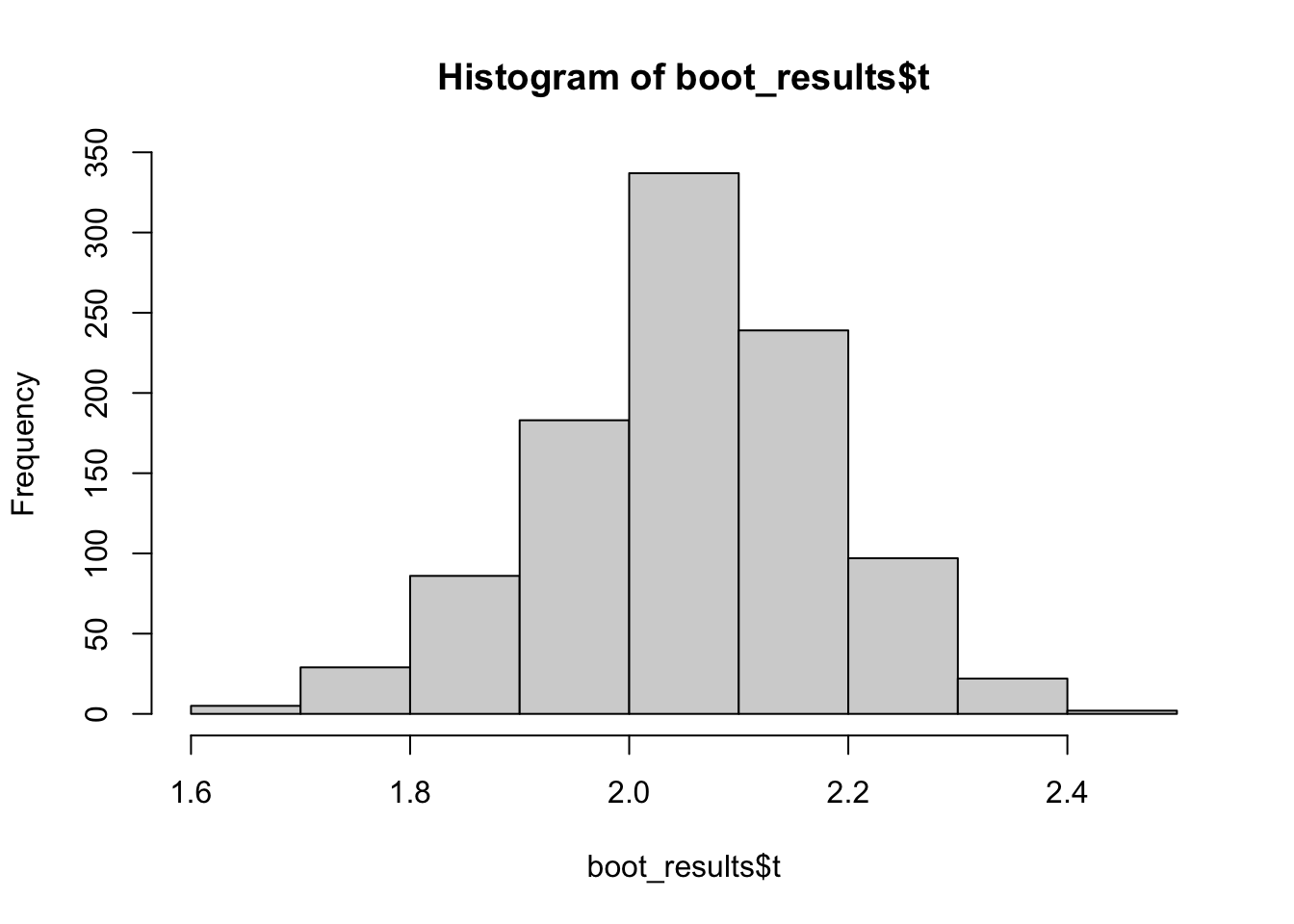
$t0 contains values of our statistic(s) in original, full, dataset:
full.fit <- lm(y~x, data = dat)
coef(full.fit)["x"] x
2.05167 boot_results$t0 # same as above x
2.05167 The Bootstrap Statistics:
originalis the same as$t0biasis a difference between the mean of bootstrap realizations (averge of$tvalues), and the value in original dataset (saved to$t0).std. erroris a standard error of bootstrap estimate, which equals standard deviation of bootstrap realizations.
sd(boot_results$t) # std.error in Bootstrap Statitiscs[1] 0.1267117Footnotes
Alternatively, you can use your one model in
train(); see https://topepo.github.io/caret/using-your-own-model-in-train.html↩︎The value of
Kmust be such that all groups are of approximately equal size. If the supplied value ofKdoes not satisfy this criterion then it will be set to the closest integer which does and a warning is generated specifying the value ofKused.↩︎We won’t discuss the second component, but in case you are interested, this is the adjusted cross-validation estimate. The adjustment is designed to compensate for the bias introduced by not using leave-one-out cross-validation.↩︎
When using
glm()with the default settings of a Gaussian family and an identity link function (the default), it performs the same operation aslm(). Thus,glm(y ~ x, data)equivalent tolm(y ~ x, data).↩︎we did not discuss this in class, but the bias tends to be downward. i.e., we tend to underestimate MSE for new data and be overly optimist.↩︎
Summary
All of the estimates summarized in the table below are estimating the same quantity: the MSE of the model when predicting new, unseen data. As discussed in class, LOOCV has lower bias but higher variance, while \(k\)-fold CV has a bit more bias but lower variance. This means that the \(k\)-fold estimate tends to be more stable (albeit slightly more biased5). As we can see, the estimates are all in the same ballpark, but they will differ notice how the \(k\)-fold runs will differ based on the random partitions into folds.
5- or 10- fold CV is often preferred in practice. It’s computationally cheaper than LOOCV and tends to perform well in most situations.