Data 311: Machine Learning
Lecture 19: Gaussian Mixture Models
University of British Columbia Okanagan
Learning Objectives
Today we will be looking at a probabilistic approach to clustering called Gaussian Mixture Models. Aims:
- Understand key components of Gaussian Mixture Models.
- Compare & contrast GMMs to other clustering techniques.
- Grasp the role of probabilistic modeling in clustering.
- Explore the Expectation-Maximization (EM) algorithm.
- Apply GMMs to a practical clustering task.
Clustering
Recall: the goal of clustering1 (a form of unsupervised learning) is to find groups such that all observations are
- more similar to observations inside their group and
- more dissimilar to observations in other groups.
Cluster analysis is widely adopted by various applications like image processing, neuroscience, economics, network communication, medicine, recommendation systems, customer segmentation, to name a few.
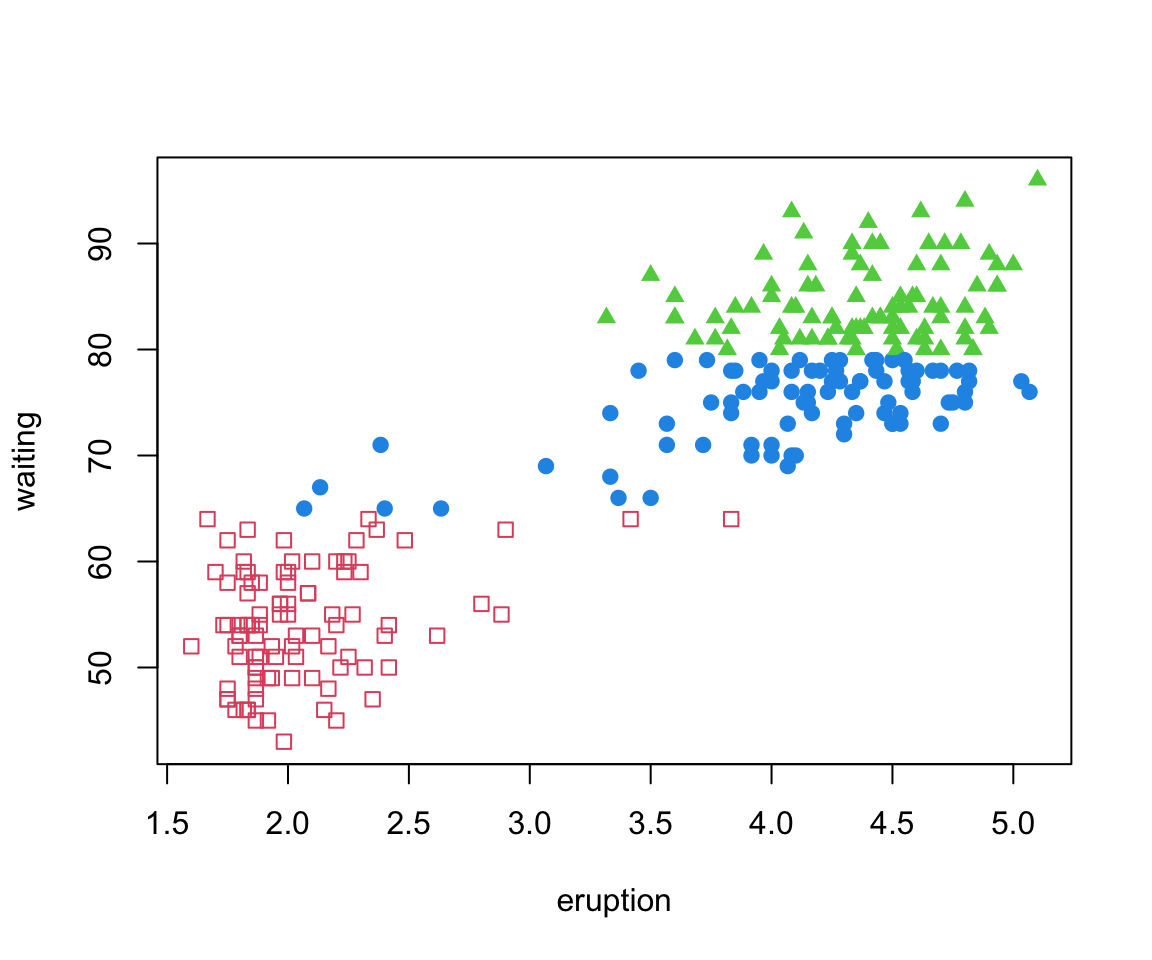
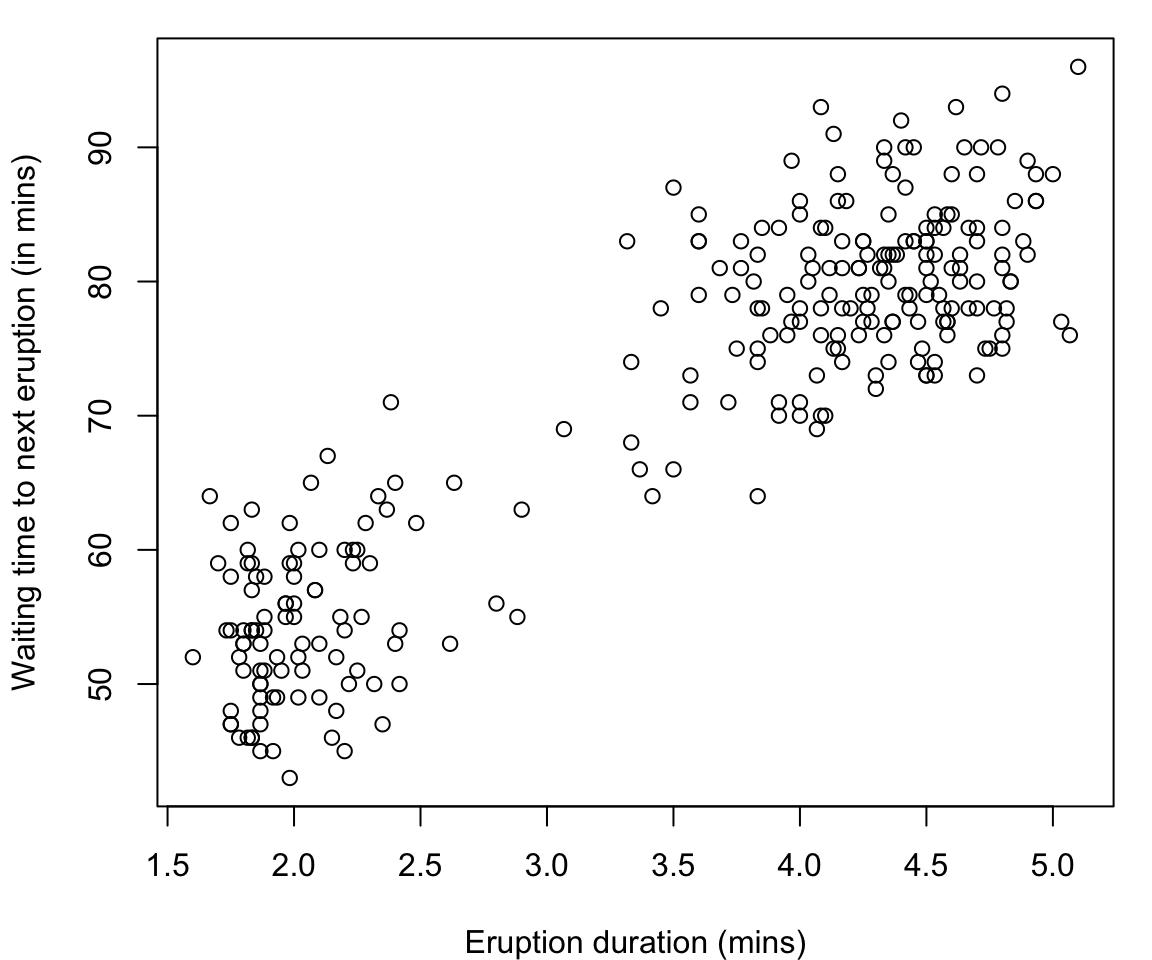
Old Faithful Data
The
faithfuldataset in R contains the measurements on time to eruption, and length of eruption time of the Old Faithful geyser in Yellowstone National Park.-
The data frame consists of 272 observations on 2 variables:
eruptionsEruption time in mins (numeric)waitingWaiting time to next eruption in mins (numeric)
N.B. to see the help file for any dataset (or function in R) use ?name_of_thing
jump to the iris data set.
Many would argue that there are two clusters:
- Short eruptions followed shortly thereafter by another eruption
- Longer eruptions with longer delay before the next eruption
Unsupervised Clustering
The geyser data falls in the unsupervised clustering problem since there are no predefined labels or outcomes.
While we can manually identify groupings (i.e. clusters) in lower dimensional space, this is impossible for higher dimensions in which we can no longer visualize our data.
Determining Clusters
Rather than creating the clusters ourselves, we look for automated ways to find these natural groupings.
Previously we saw how k-means and hierarchical clustering can cluster this data
We used “elbow method” for kmeans and large vertical gaps in the dendrogram to determine the number of clusters.
Another option could be running
Mclust* to obtain a optimal* partition of the data.
faithful clustered
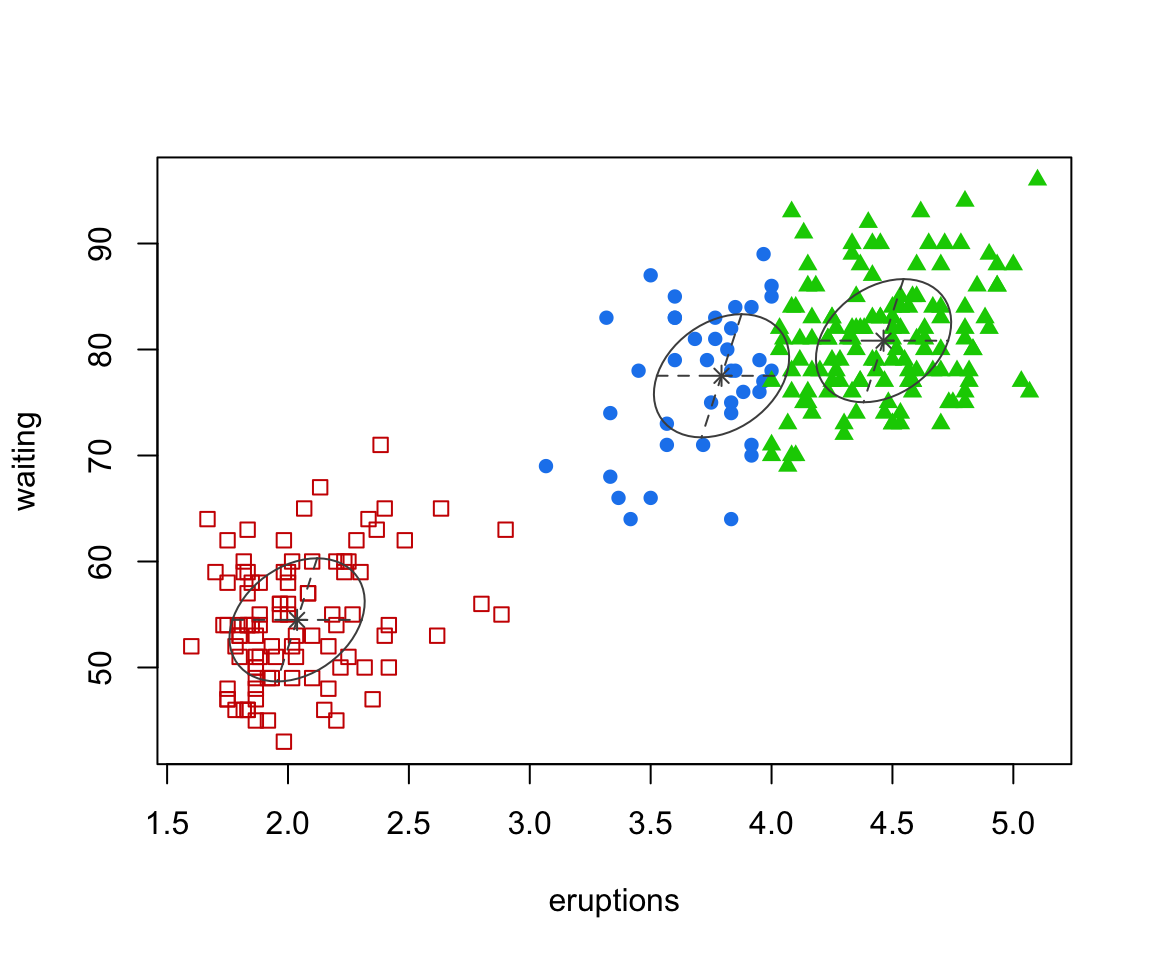
The points are coloured by group allocation.
Interestingly, mclust determines that there are 3 groups in the data.
But how did mclust determine that 3 clusters was optimal?
Recall Kmeans
Code
# Load the data
library(MASS)
# Define a function to calculate total within-cluster sum of squares (WSS)
wss <- function(data, k) {
kmeans(data, centers = k, nstart = 25)$tot.withinss
}
# Compute WSS for k = 1 to 10 clusters
set.seed(42) # For reproducibility
k_values <- 1:10
wss_values <- sapply(k_values, wss, data = faithful)
# Plot the scree plot
plot(k_values, wss_values, type = "b", pch = 19, frame = FALSE,
xlab = "Number of Clusters (k)",
ylab = "Total Within-Cluster Sum of Squares (WSS)",
main = "Scree Plot for K-Means Clustering")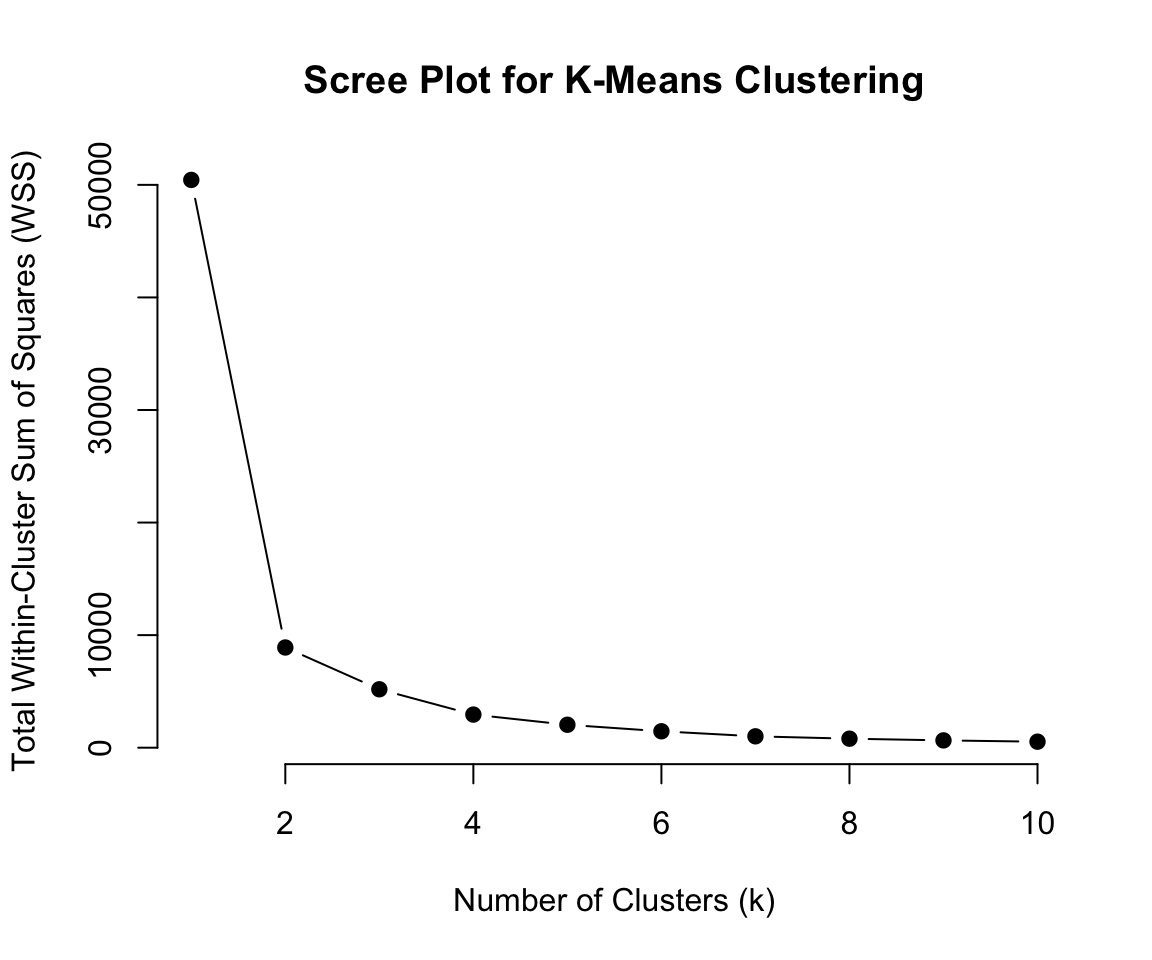
Recall Kmeans
- Partitions data into \(k\) clusters by minimizing within-cluster variance.
- Assumes spherical clusters and hard assignments.
Limitations:
- Assumes clusters are similar in size and shape.
- Cannot model overlapping clusters.
GMM addresses these limitations with a probabilistic approach.
From k-Means to GMM
Kmeans
centroid-based clustering algorithms
“hard”1 clustering
assumes spherical, equal-sized clusters
GMM
Distribution-based clustering
soft2 clustering
supports ellipsoidal clusters and varying sizes
Mixture Models
Resources
[MP] Mixture Model-Based Classification by Paul D. McNicholas
[FS] Finite Mixture and Markov Switching Models by Fruhwirth-Schnatter, S. (2006)
[MG] Finite Mixture Models by McLachlan, Geoffrey J; Peel, David/ New York: John Wiley & Sons
[LB] Mixture models: theory, geometry, and applications by Lindsay, Bruce G
Mixture Models
Mixture models are just a convex combination of statistical distributions. In general, for a random vector \(\mathbf{X}\), a mixture distribution is defined by \[f(\mathbf{x}) = \sum_{g=1}^G \pi_g p_g(\mathbf{x} \mid \theta_g)\] where \(G\) is the number of groups1, \(0 \leq \pi_g \leq 1\) and \(\sum_{g=1}^G \pi_g = 1\) and \(p_g(\mathbf{x} \mid \theta_g)\) are statistical distributions with parameters \(\theta_g\).
Terminology
The \(\pi_g\) are called mixing proportions and we will write \(\mathbf{\pi} = (\pi_1, \pi_2, \dots, \pi_G)\)
The \(p_g(\mathbf{x})\) are called component densities
Since the \(p_g(\mathbf{x})\) are densities, \(f(\mathbf{x})\) describes a density called a \(G\)-component finite mixture density.
Statistical Distributions
In theory, \(p_g(\mathbf{x} \mid \theta_g)\) could be a different distribution type for each \(g\).
In practice, however, \(p_g(\mathbf{x} \mid \theta_g)\) is assumed to be the same across groups, but with different \(\theta_g\).
Most commonly, we assume a a Gaussian mixture model (GMM) where \(p_g(\mathbf{x} \mid \theta_g)\) is assumed to be multivariate Gaussian (aka multivariate Normal), denoted \(\phi(\mathbf{x} | \mu_g, \mathbf{\Sigma}_g)\)
Gaussian Mixture model
Thus \(f(\mathbf{x})\) — sometimes written \(f(\mathbf{x} | \Theta)\) or \(f(\mathbf{x} ; \Theta)\) — is a Gaussian mixture model would have the form: \[\begin{align} f(\mathbf{x} | \Theta) &= \sum_{g=1}^G \pi_g \phi(\mathbf{x} \mid \boldsymbol\mu_g, \mathbf{\Sigma}_g) \\ &= \sum_{g=1}^G \pi_g \frac{|\mathbf{\Sigma}|^{-1/2}}{(2\pi)^{k/2}} \exp \left( - \frac{1}{2} (\mathbf{x} - \boldsymbol{\mu}_g)^{\mathrm {T}} \mathbf{\Sigma}^{-1} (\mathbf{x} - \boldsymbol{\mu}_g) \right) \end{align}\] where \(\phi\) denote the normal distribution, \(\boldsymbol{\mu}_g\) is the mean vector for group \(g\) and \(\mathbf{\Sigma}_g\) is the covariance matrix for group \(g\), and \(\Theta = \{\boldsymbol{\mu}_1, ..., \boldsymbol{\mu}_G, \mathbf{\Sigma}_1, ..., \mathbf{\Sigma}_G, \pi_1, ..., \pi_G\}\)
Components of GMM
Means (\(\mu_k\)): Cluster centers.
Covariances (\(\Sigma_k\)): Shape and spread of clusters.
Mixing Coefficients (\(\pi_k\)): Proportion of data in each cluster.
Distribution Alternatives
Other robust alternatives include:
Mixtures of Student-\(t\) distributions1 (heavier tails)
-
Mixtures of skewed distributions
- skew-normal (Lin et al, 2007)
- skewed-\(t\) Vrbik and McNicholas, 2012
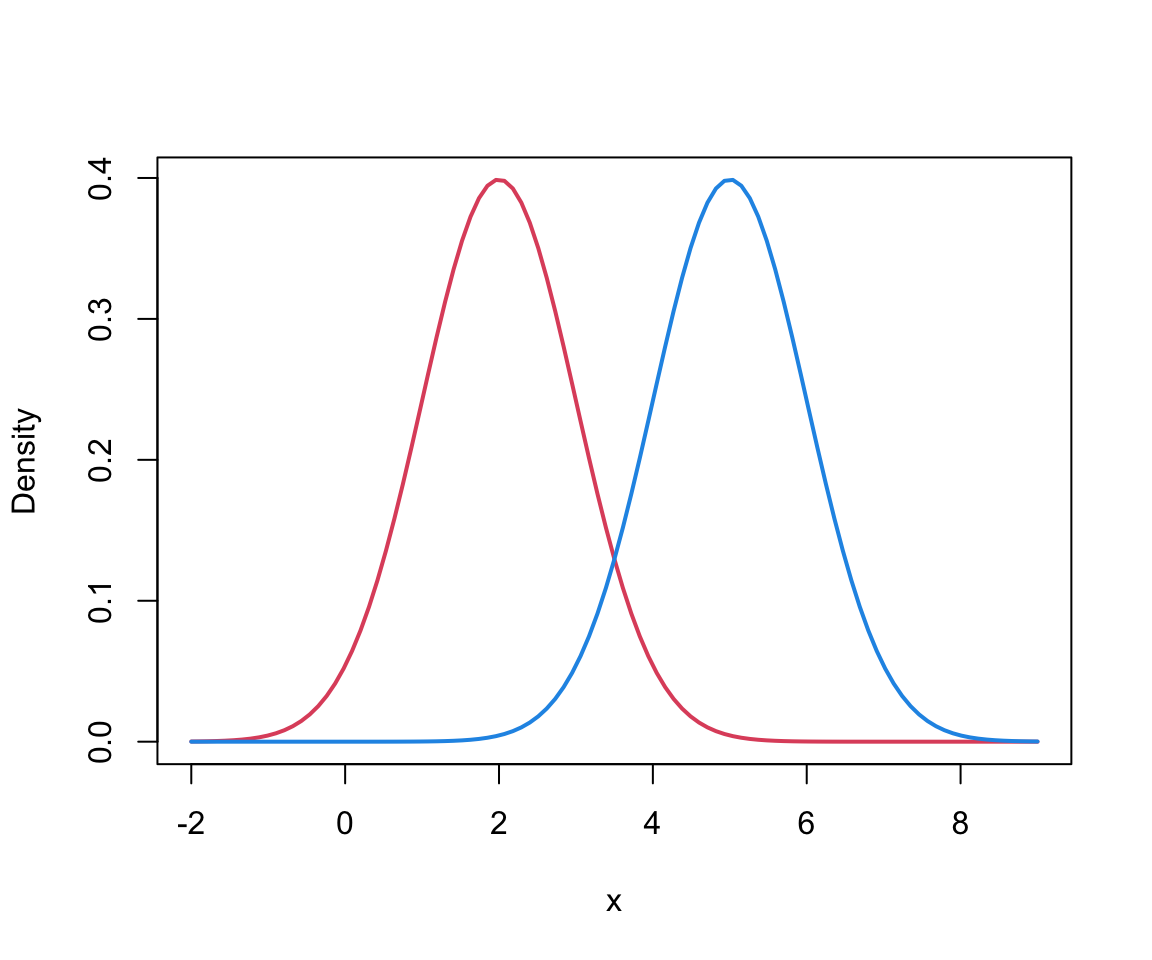
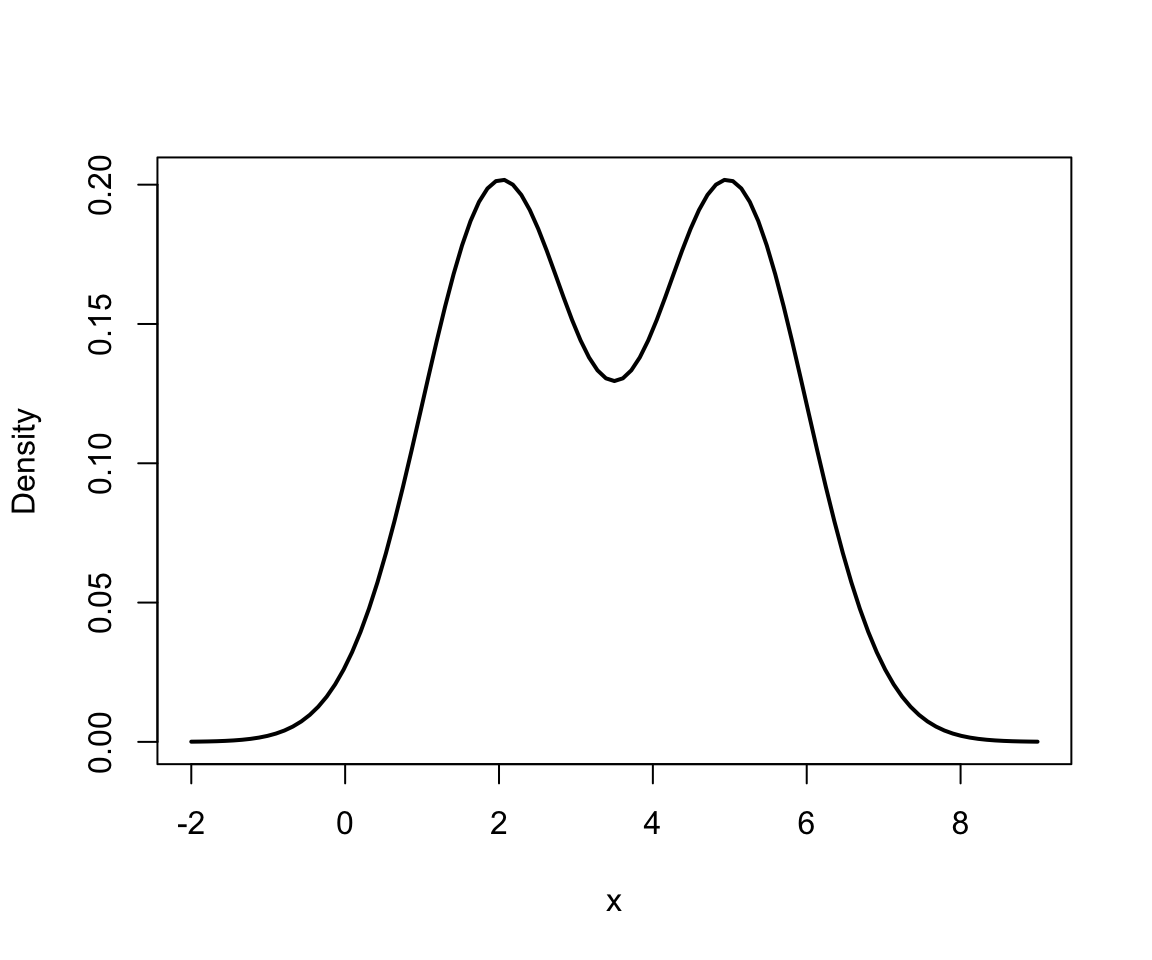
Finite mixture in 1D
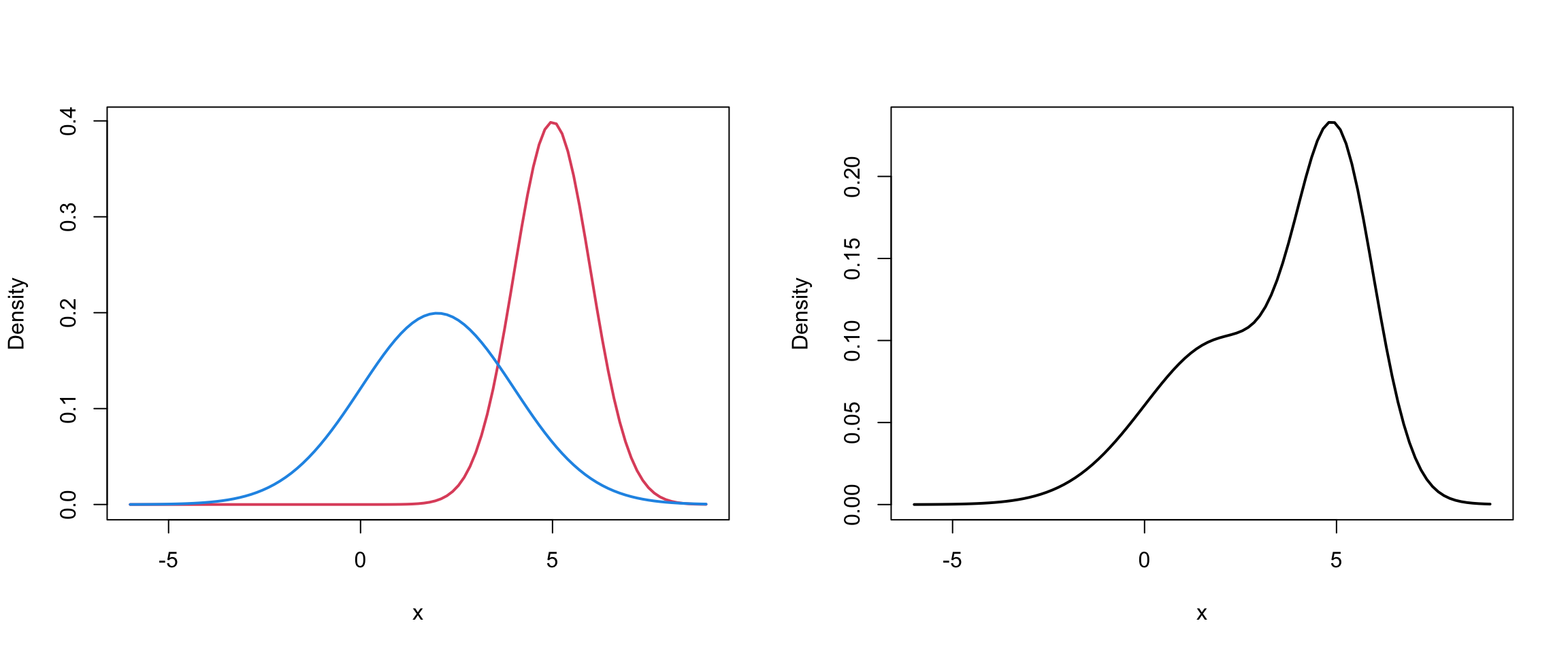
This can be viewed as a mixture of 2 univariate Gaussians….
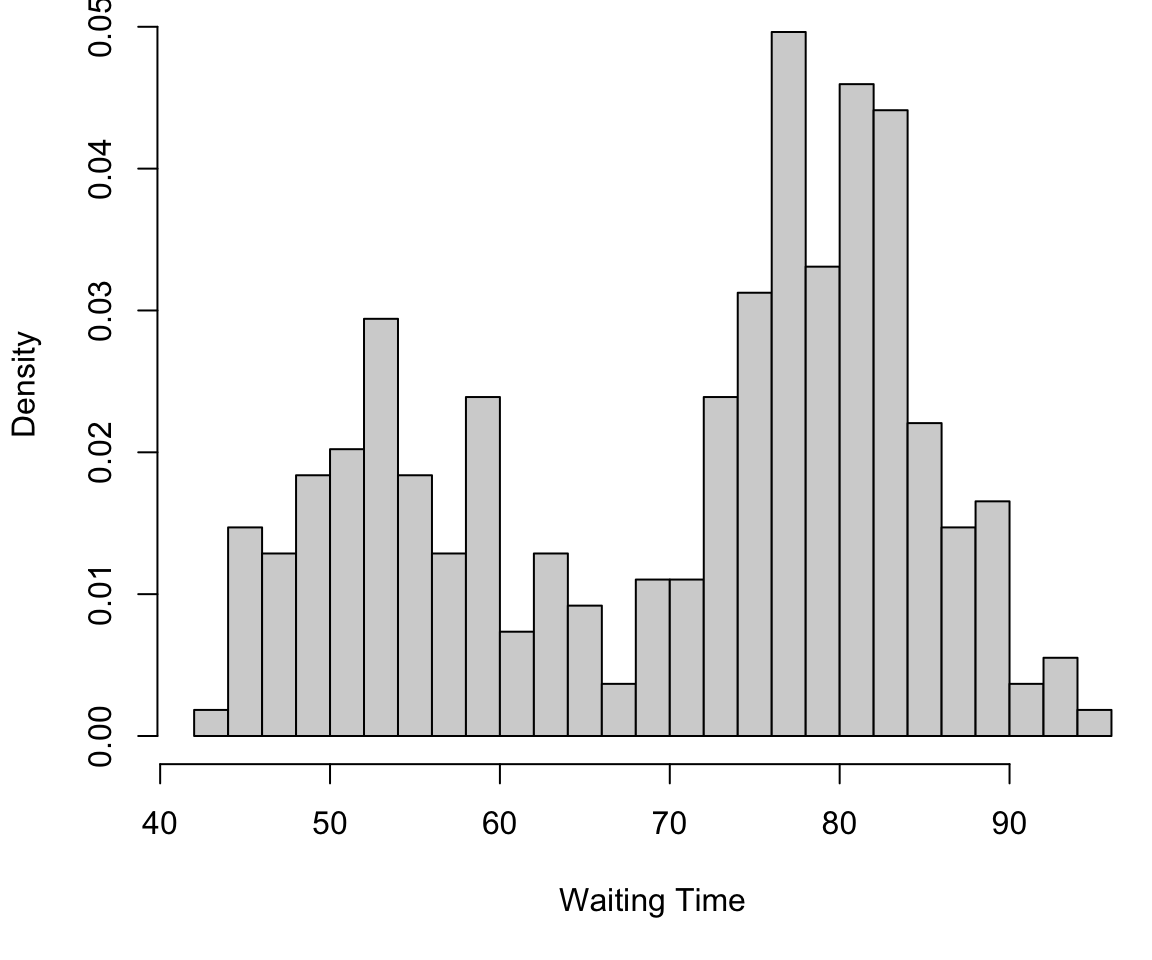
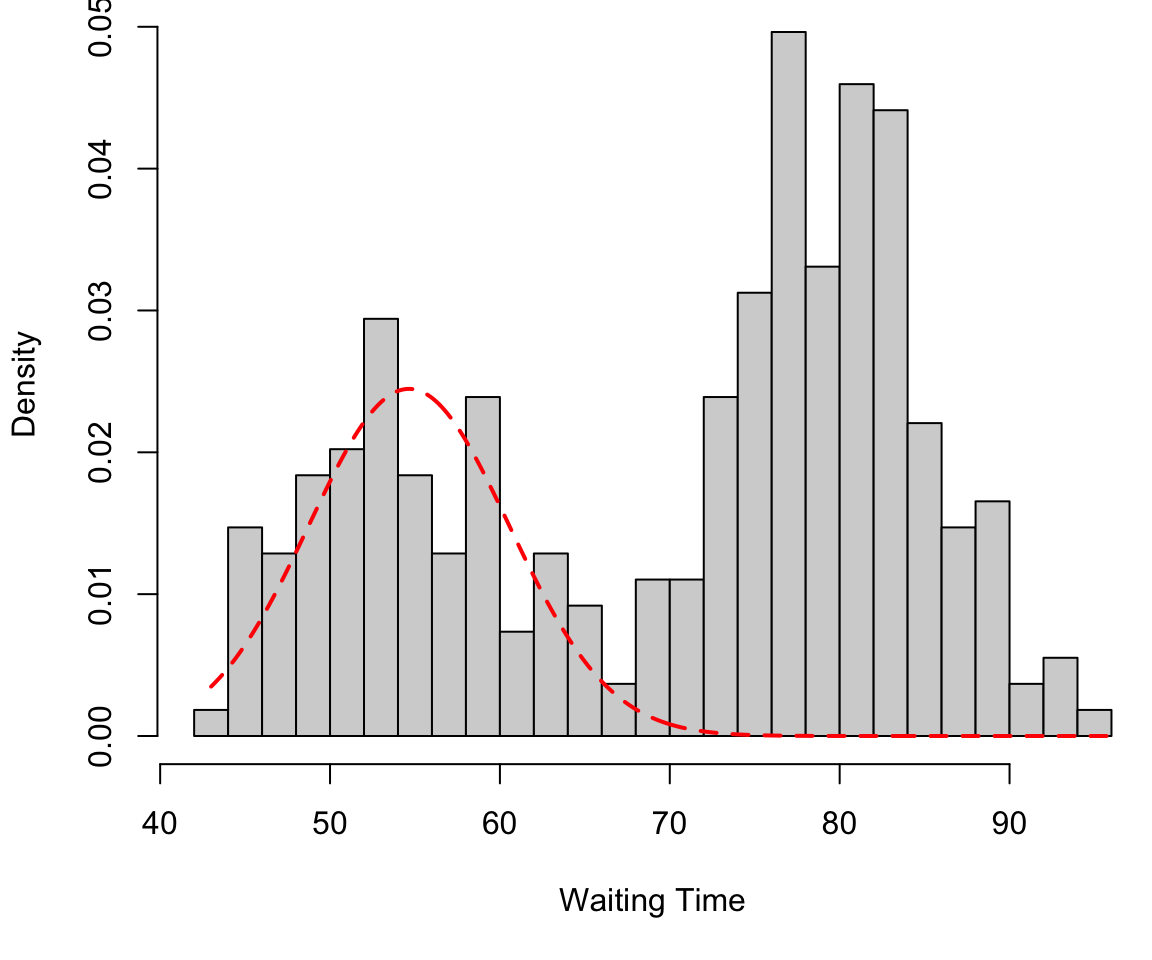
Finite mixture in 1D
Component 1 (in red) \[f_1 \sim N(\mu = 54.6, \sigma = 5.9)\]
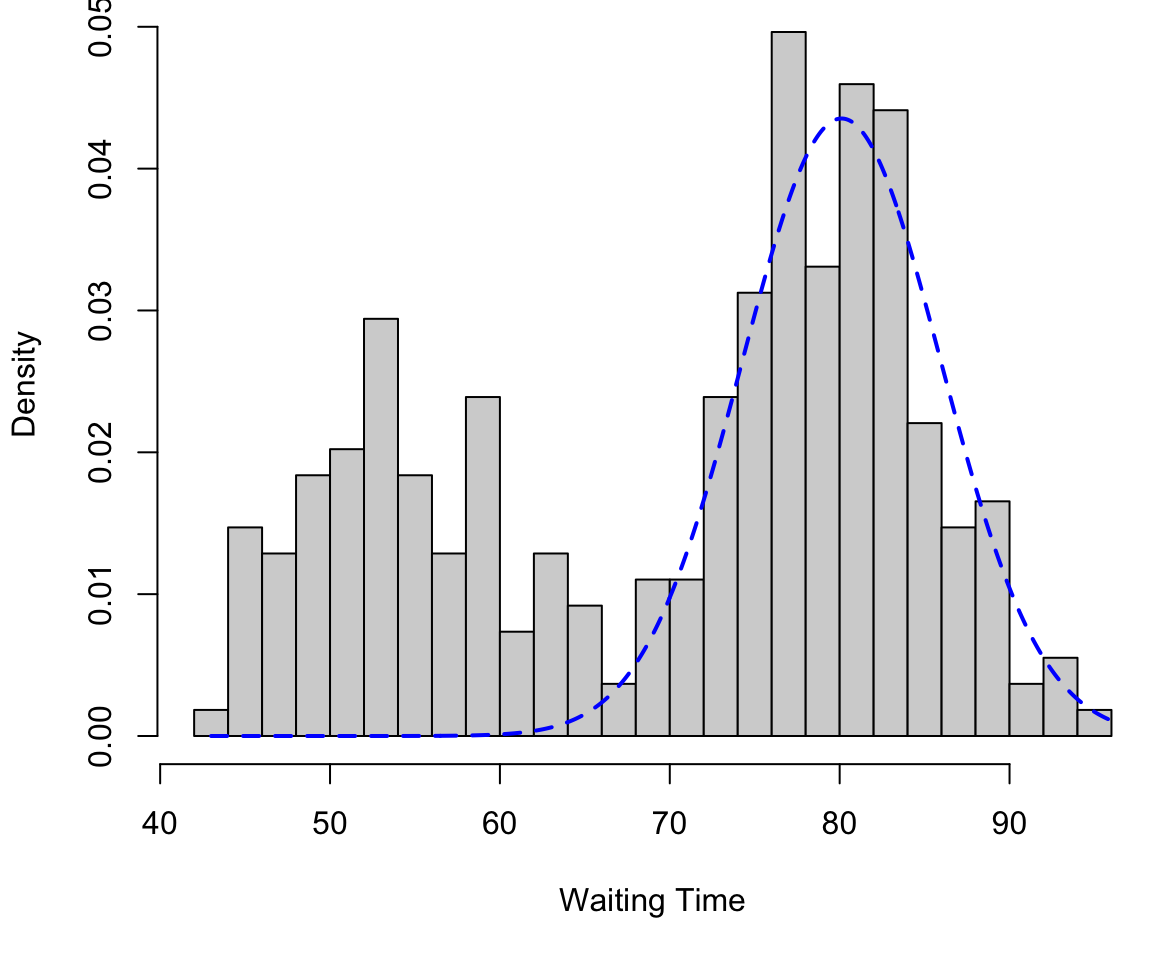
Finite mixture in 1D
Component 2 (in blue) \[f_2 \sim N(\mu = 80.1, \sigma = 5.8)\]
Finite mixture in 1D
And mixture (in black): \[ 0.4 f_1 + 0.6f_2 \]
Code
# Load required libraries
library(mclust)
# Load the faithful dataset
data("faithful")
waiting <- faithful$waiting
# Fit a Gaussian Mixture Model with 2 components
set.seed(42)
gmm <- Mclust(waiting, G = 2, modelNames = "V")
# Extract GMM parameters
means <- gmm$parameters$mean
sds <- sqrt(gmm$parameters$variance$sigmasq)
proportions <- gmm$parameters$pro
# Create a sequence for density plotting
x_vals <- seq(min(waiting), max(waiting), length.out = 500)
# Compute individual and overall densities
density1 <- proportions[1] * dnorm(x_vals, mean = means[1], sd = sds[1])
density2 <- proportions[2] * dnorm(x_vals, mean = means[2], sd = sds[2])
overall_density <- density1 + density2
par(mar=c(5,4,0,0) +0.1)
# Plot the histogram and density curves
hist(waiting, freq = FALSE, breaks = 20, main = "",
xlab = "Waiting Time")
lines(x_vals, overall_density, col = "black", lwd = 2, lty = 1) # Overall density
lines(x_vals, density1, col = "red", lwd = 2, lty = 2) # Component 1
lines(x_vals, density2, col = "blue", lwd = 2, lty = 2) # Component 2
legend("topleft", legend = c("Overall Density", "Component 1", "Component 2"),
col = c("black", "red", "blue"), lwd = 2, lty = c(1, 2, 2), bty = "n")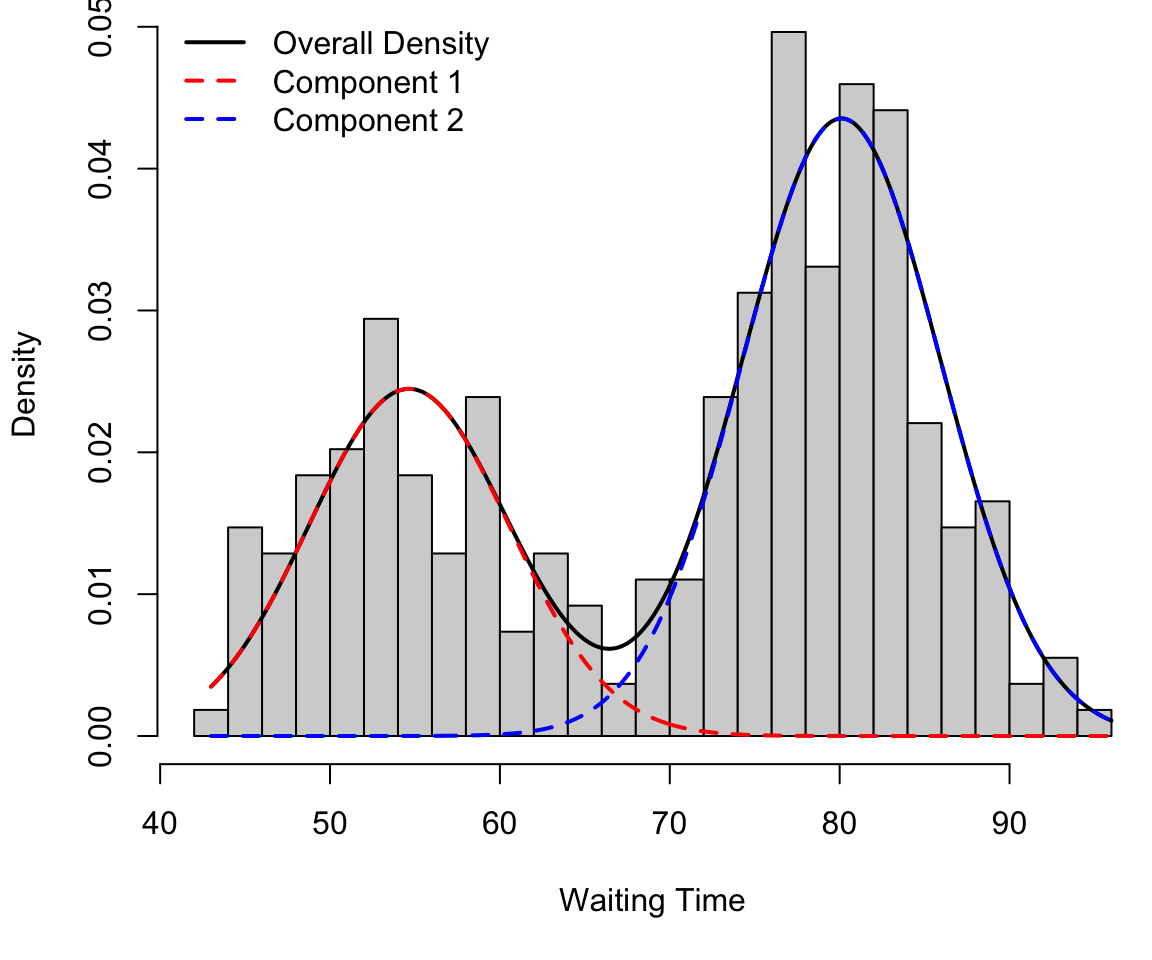
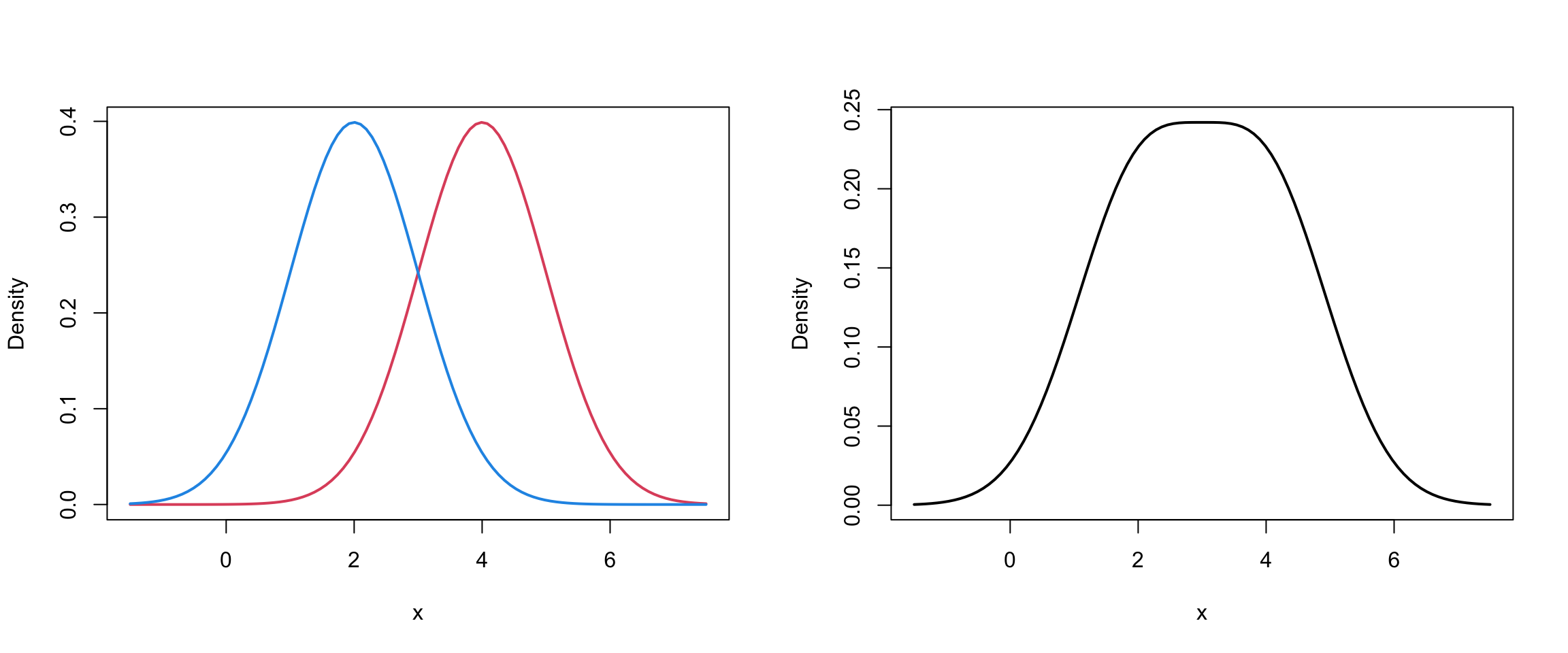
Two Univariate Normals I
Mixture of 2 univariate Normals II
Mixture of 2 univariate Normals II
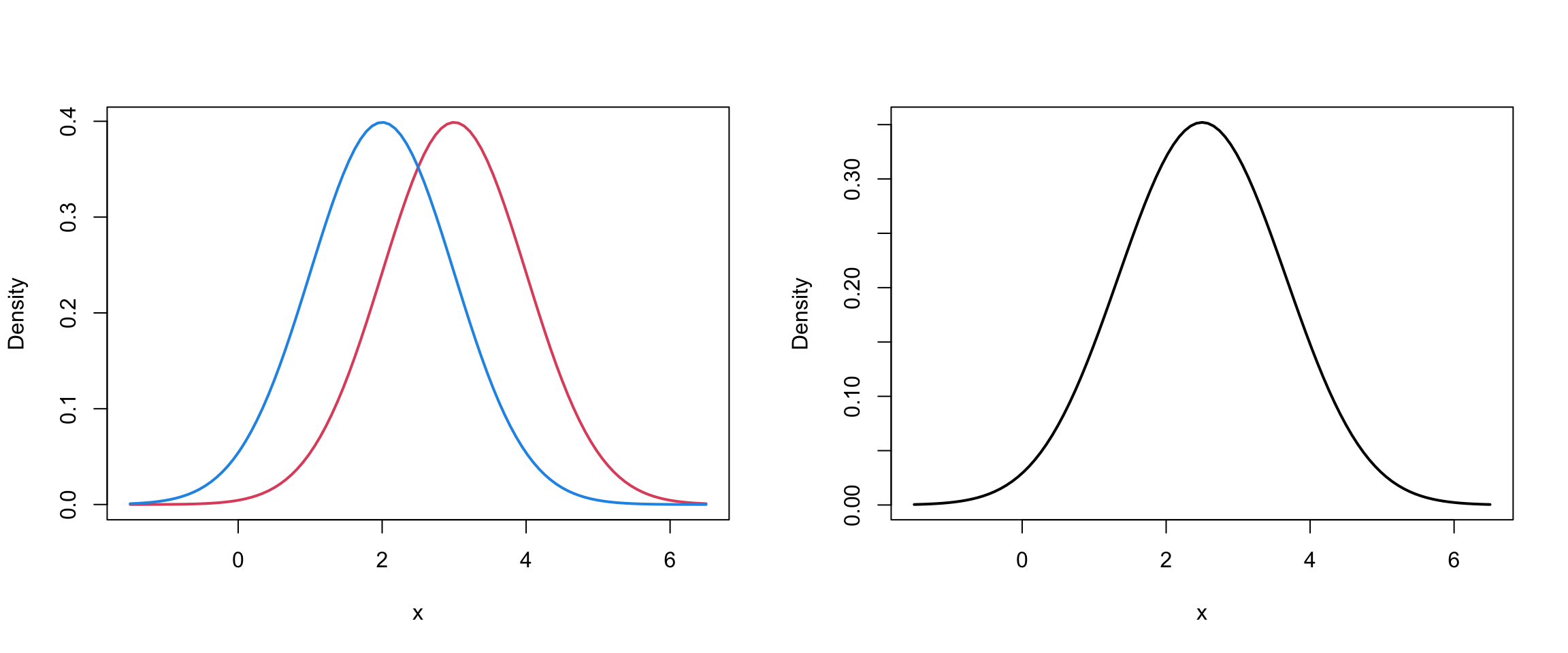
Mixture of 2 univariate Normals III
Identifiability I
Inference for finite mixture modles is typical in that we want to estimate parameters \(\Theta = \{\theta_1, ... , \theta_G, \pi_1, ...,\pi_G\}\)
However, this estimation is only meaningful if \(\Theta\) is identifiable.
In general, a family of densities \(\{ p(\mathbf{x} | \Theta) : \Theta \in \Omega \}\) is identifiable if: \[p(\mathbf{x} | \Theta) = p(\mathbf{x} |\Theta^*) \iff \Theta^* = \Theta\] In the case of mixture models this gets a bit complicated…
Identifiability II
One problem is that component labels may be permuted:
\[\begin{align} p(x | \Theta) &= \pi_1 p(x | \mu_1, \sigma_1^2) + \pi_2 p(x | \mu_2, \sigma_2^2)\\ p(x | \Theta^*) &=\pi_2 p(x | \mu_2, \sigma_2^2) + \pi_1 p(x | \mu_1, \sigma_1^2) \end{align}\]
The identifiable condition is not satisfied because the component labels can be interchanged yet \(p(x | \Theta) = p(x | \Theta^*)\)
As [MG] point out, even though this class of mixtures may be identifiable, \(\Theta\) is not.
Identifiability III
The identifiability issue that arises due to the interchanging of component labels can be handled by imposing appropriate constraints on \(\Theta\)
For example, the \(\pi_g\) could be constrained so that \(\pi_i \geq \pi_k\) for \(i>k\).
Also we can avoid empty components by insisting that \(\pi_g > 0\) for all \(g\).
Mixture Models for clustering
Finite mixture models are ideally suited for clustering
Clusters correspond to the \(1,...,g,...G\) sub-populations/groups/components in a finite mixtures model.
So, let’s attempt the to derive the MLE of the Gaussian mixture model …
The Likelihood
The likelihood function for this mixture is \[\begin{align} L(\Theta) &= f(\mathbf{x} | \Theta) = \prod_{i=1}^n \left[ \sum_{g=1}^G \pi_g \phi(\mathbf{x} \mid \boldsymbol\mu_g, \mathbf{\Sigma}_g) \right] \end{align}\] and the so-called log-likelihood is: \[\begin{align} \ell(\Theta) &= \log(L(\Theta)) = \sum_{i=1}^N \log\left[ \sum_{g=1}^G \pi_g \phi(\mathbf{x} \mid \boldsymbol\mu_g, \mathbf{\Sigma}_g) \right] \end{align}\]
If you try to differentiate w.r.t. to \(\mu_g\), set =0, and solve, you would find that this has no analytic solution.
EM algorithm
Most commonly, we use the expectation-maximization (EM) algorithm, which helps us find parameter estimates with missing data (group memberships are missing).
More generally, the EM is an iterative algorithm for finding maximum likelihood parameters in statistical models, where the model depends on unobserved latent variables.
We assume that each observed data point has a corresponding unobserved latent variable specifying the mixture component to which each data point belongs.
Latent Variable
We introduce a new latent1 variable that represents the group memberships: \[\begin{equation} Z_{ig} = \begin{cases} 1 & \text{if } \mathbf{x}_i \text{ belongs to group }g \\ 0 & \text{otherwise} \end{cases} \end{equation}\]
We call \(\{X, Z\}\) our “complete” data.
Now that we have that, let’s look at a non-technical summary of the EM algorithm…
Latent Variable Representation
The joint probability of a data point \(x_i\) and its latent variable \(Z_{ig}\) is:
\[ p(x_i, Z_{ig} \mid \Theta) = p(Z_{ig} \mid \Theta) p(x_i \mid Z_{ig}, \Theta) \]
The complete likelihood assumes we know the latent cluster assignments:
\[ \begin{align*} L_c(\Theta) &= \prod_{i=1}^n \prod_{g=1}^G \left[\pi_g \phi(x_i|\mu_g, \sigma_g)\right]^{\hat{z}_{ig}}\\ \ell_c(\Theta) &= \sum_{i=1}^n \sum_{g=1}^G \hat{z}_{ig} \left[ \log (\pi_g) + \log (\phi(x_i|\mu_g, \sigma_g) )\right] \end{align*} \]
EM Pseudocode
- Start the algorithm with random values for \(\hat z_{ig}\).
- Mstep: Assuming those \(\hat{z}_{ig}\) are correct, find the maximum likelihood estimates (MLE)
- Estep: Assuming those parameters are correct, find the expected value of group memberships \[\hat{z}_{ig} = \frac{\pi_g \phi(\mathbf{x} \mid \boldsymbol{\mu}_g, \mathbf{\Sigma}_g) }{\sum_{g=1}^G \pi_g \phi(\mathbf{x} \mid \boldsymbol{\mu}_g, \mathbf{\Sigma}_g)}\]
- Repeat 2. and 3. until “changes” are minimal.
Mstep
For the “M-step”, i.e. maximization step, and given the \(z\)’s calculated in the E-step …
\[ \begin{align*} \mu_g &= \frac{\sum_{i=1}^n z_{ig} x_i}{\sum_{i=1}^n z_{ig}} & \pi_g &= \frac{\sum_{i=1}^n z_{ig}}{n} \end{align*} \] \[ \Sigma_g = \frac{\sum_{i=1}^n z_{ig} (x_i - \mu_g)(x_i - \mu_g)^T}{\sum_{i=1}^N r_{ig}} \]
where \(n\) is the total number of data points.
Fuzzy z’s
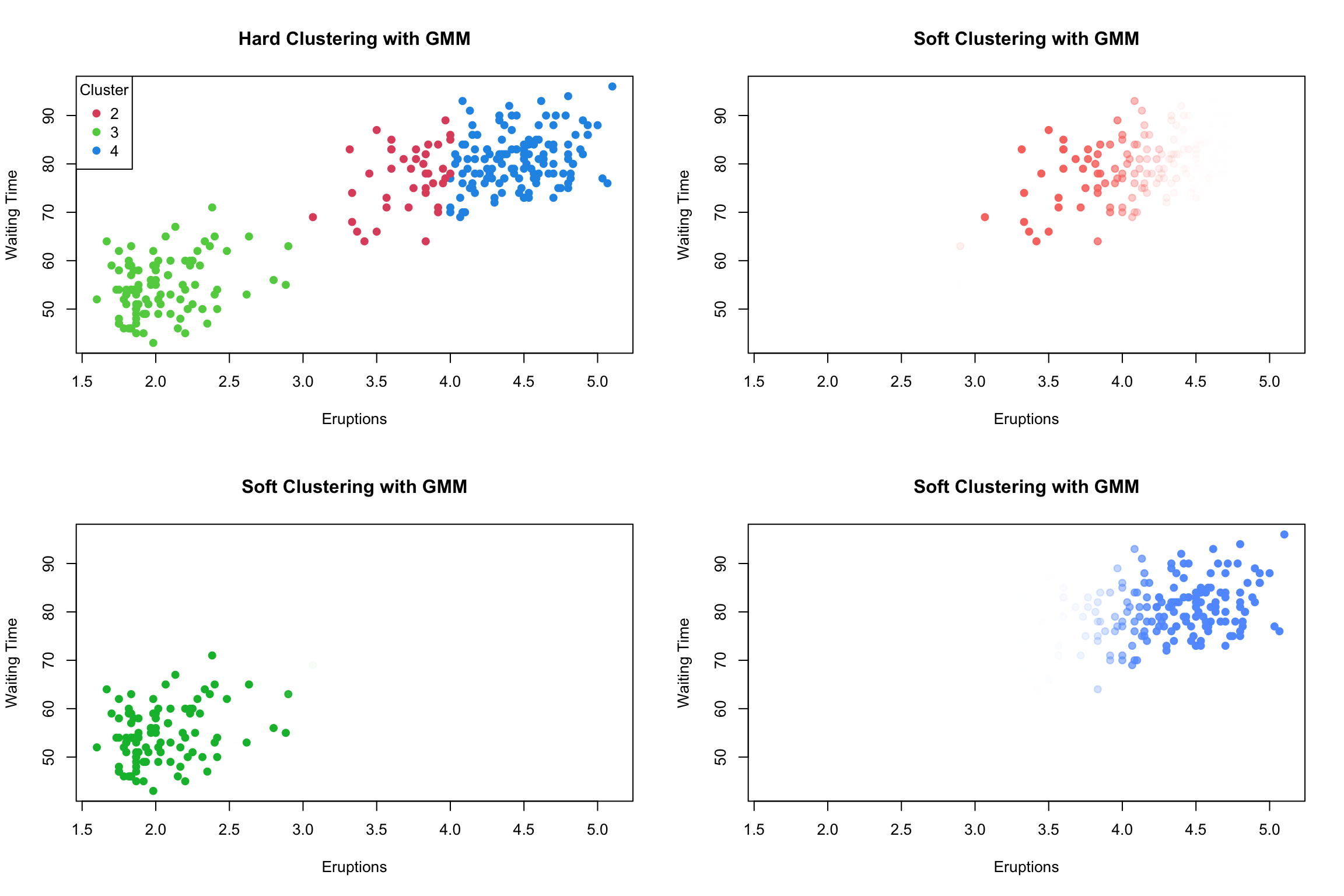
The resulting \(\hat{z}_{ig}\) are not going to be 0’s and 1’s when the algorithm converges.
For each observation \(\mathbf{x}_i\) we end up with vector of probabilities (\(\hat{z}_{i1}\),…,\(\hat{z}_{iG}\)) which we denote by \(\hat{\mathbf{z}}_i\)
To convert this to a “hard” clustering, we simply assign the observation to \(g* = \arg \max_{g} \hat z_{ig}\)
e.g. for \(\mathbf{x}_i\) with \(\hat{\mathbf{z}}_i = (0.25, 0.60, 0.15)\) –> in this case \(\mathbf{x}_i\) would get assigned to group 2
MCLUST example
To see this in action, lets take a closer look at the MCLUST output when it was fit to our old faithful geyser data.
To recap:
MCLUST imposes the GMM structure.
We introduce indicator \(z_{ig}\) such that \(z_{ig} = 1\) is observation \(i\) belongs to group \(g\) and \(z_{ig}= 0\) otherwise.
The model is fitted using the EM alogrithm
Classification vector
Our estimates of \(z_{ig}\), denoted \(\hat z_{ig}\) determine our clusters: \[\begin{equation} \text{MAP}{\hat{z}_{ig}} = \begin{cases} 1 & \text{if } \max_g \{\hat z_{ig}\} \\ 0 & \text{otherwise} \end{cases} \end{equation}\]
[,1] [,2]
[1,] 1.030748e-04 0.9998969252
[2,] 9.999098e-01 0.0000901813
[3,] 4.146885e-03 0.9958531153
[4,] 9.675687e-01 0.0324313138
[5,] 1.217871e-06 0.9999987821
[6,] 9.998111e-01 0.0001889468 [,1] [,2]
[1,] 0 1
[2,] 1 0
[3,] 0 1
[4,] 1 0
[5,] 0 1
[6,] 1 0[1] 2 1 2 1 2 1Soft Clustering
GMM is a type of soft clustering.
This means that for data object can exist in more than one cluster with a certain probability or degree of membership.
This is unlike hard clustering methods which define a non-overlapping partition of the data set (eg. kmeans)
The matrix of \(\hat z_{ig}\) is often referred to has a “fuzzy” z matrix.
The rows of this matrix should necessarily sum to 0.
iClicker
What role do the mixing coefficients (\(\pi_g\)) play in GMM?
- They determine the variance of each Gaussian component.
- They define the mean of each Gaussian component.
- They represent the proportion of data points expected to belong to each cluster.
- They are used to compute the covariance matrix.
Problem 1
EM algorithm is a deterministic, monotonic optimization algorithm; however, it is susceptible to local maxima!
Hence if we begin the algorithm at different starting points, we risk getting different clustering results.

Image Source: CLP-1
Smart/multiple starts
One solution to problem 1 is is start your algorithm and several different initialization
Another potential solution it to start in a smart location (to be determined by some other clustering method, eg. kmeans)
Problem 2
The total number of “free” parameters in a GMM is: \[\begin{equation} (G-1) + Gp + Gp(p+1)/2 \end{equation}\]
\(Gp(p+1)/2\) of these are from the group covariance matrices \(\mathbf{\Sigma}_1,..., \mathbf{\Sigma}_G\)
For the EM, we have to invert these \(G\) covariance matrices for each iteration of the algorithm.
This can get quite computationally expensive…
Common Covariance
One potential solution to problem 2 is to assumes that the covariance are equal across groups
The constraint of \(\mathbf{\Sigma}_g = \mathbf{\Sigma}\) for all \(g\) this reduces to \(p(p+1)/2\) free parameters in the common covariance matrix.
This however is a very strong assumption about the underlying data, so perhaps we could impose a softer constraint …
Towards MCLUST
MCLUST exploits an eigenvalue decomposition of the group covariance matrices to give a wide range of structures that uses between 1 and \(Gp(p+1)/2\) free parameters.
The eigenvalue decomposition has the form \[\begin{equation} \mathbf{\Sigma}_g = \lambda_g \mathbf{D}_g \mathbf{A}_g \mathbf{D}'_g \end{equation}\] where \(\lambda_g\) is a constant, \(\mathbf{D}_g\) is a matrix consisting of the eigenvectors of \(\mathbf{\Sigma}_g\) and \(\mathbf{A}_g\) is a diagonal matrix with entries proppotional the the eigenvalues of \(\mathbf{\Sigma}_g\).
Constraints & MCLUST
We may constrain the components of the eigenvalue decomposition of \(\mathbf{\Sigma}_g\) across groups of the mixture model
Furthermore, the matrices \(\mathbf{D}_g\) and \(\mathbf{A}_g\) may be set to the identity matrix \(\mathbf{I}\)
Fraley & Ratery (2000) describe the
mclustsoftware, which is available as an R package that efficiently performs this constrained model-based clustering.
MCLUST models
| ID | Volume | Shape | Orientation | Covariance Decomposition | Number of free Covariance parameters |
|---|---|---|---|---|---|
| EII | E | S | \(\lambda \mathbf{I}\) | 1 | |
| VII | V | S | \(\lambda_g \mathbf{I}\) | \(G\) | |
| EEI | E | E | AA | \(\lambda \mathbf{A}\) | \(p\) |
| VEI | V | E | AA | \(\lambda_g \mathbf{A}\) | \(p+G-1\) |
| EVI | E | V | AA | \(\lambda \mathbf{A}_g\) | \(pG-G+1\) |
| VVI | V | V | AA | \(\lambda_g \mathbf{A}_g\) | \(pG\) |
| EEE | E | E | E | \(\lambda\mathbf{D}\mathbf{A}\mathbf{D}'\) | \(p(p+1)/2\) |
| EEV | E | E | V | \(\lambda\mathbf{D}_g\mathbf{A}\mathbf{D}'_g\) | \(Gp(p+1)/2-(G-1)p\) |
| VEV | V | E | V | \(\lambda_g\mathbf{D}_g\mathbf{A}\mathbf{D}'_g\) | \(Gp(p+1)/2-(G-1)(p-1)\) |
| VVV | V | V | V | \(\lambda_g\mathbf{D}_g\mathbf{A}_g\mathbf{D}'_g\) | \(Gp(p+1)/2\) |
MCLUST Cluster Shapes

Cluster shapes that correspond to the covariance structures on the previous slide
| Model | Σk | Distribution | Volume | Shape | Orientation |
|---|---|---|---|---|---|
| EII | λI | Spherical | Equal | Equal | — |
| VII | λkI | Spherical | Variable | Equal | — |
| EEI | λA | Diagonal | Equal | Equal | Coordinate axes |
| VEI | λkA | Diagonal | Variable | Equal | Coordinate axes |
| EVI | λAk | Diagonal | Equal | Variable | Coordinate axes |
| VVI | λkAk | Diagonal | Variable | Variable | Coordinate axes |
| EEE | λDAD⊤ | Ellipsoidal | Equal | Equal | Equal |
| EVE | λDAkD⊤ | Ellipsoidal | Equal | Variable | Equal |
| VEE | λkDAD⊤ | Ellipsoidal | Variable | Equal | Equal |
| VVE | λkDAkD⊤ | Ellipsoidal | Variable | Variable | Equal |
| EEV | 𝜆𝑫𝑘𝑨𝑫⊤𝑘 | Ellipsoidal | Equal | Equal | Variable |
| VEV | 𝜆𝑘𝑫𝑘𝑨𝑫⊤𝑘 | Ellipsoidal | Variable | Equal | Variable |
| EVV | 𝜆𝑫𝑘𝑨𝑘𝑫⊤𝑘 | Ellipsoidal | Equal | Variable | Variable |
| VVV | 𝜆𝑘𝑫𝑘𝑨𝑘𝑫⊤𝑘 | Ellipsoidal | Variable | Variable | Variable |
MCLUST Cluster Shapes
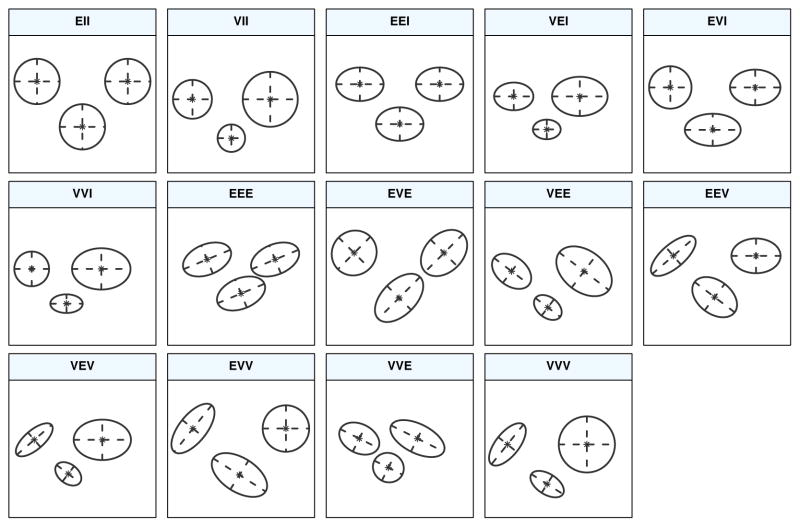
Cluster shapes that correspond to the covariance structures on the previous slide [source]
faithful finite mixture 2D
----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust EEE (ellipsoidal, equal volume, shape and orientation) model with 3
components:
log-likelihood n df BIC ICL
-1126.326 272 11 -2314.316 -2357.824
Clustering table:
1 2 3
40 97 135
Mixing probabilities:
1 2 3
0.1656784 0.3563696 0.4779520
Means:
[,1] [,2] [,3]
eruptions 3.793066 2.037596 4.463245
waiting 77.521051 54.491158 80.833439
Variances:
[,,1]
eruptions waiting
eruptions 0.07825448 0.4801979
waiting 0.48019785 33.7671464
[,,2]
eruptions waiting
eruptions 0.07825448 0.4801979
waiting 0.48019785 33.7671464
[,,3]
eruptions waiting
eruptions 0.07825448 0.4801979
waiting 0.48019785 33.7671464The “best” model is 3 components
This assumes the EEE covariance structure
But how was this determined?
BIC plot
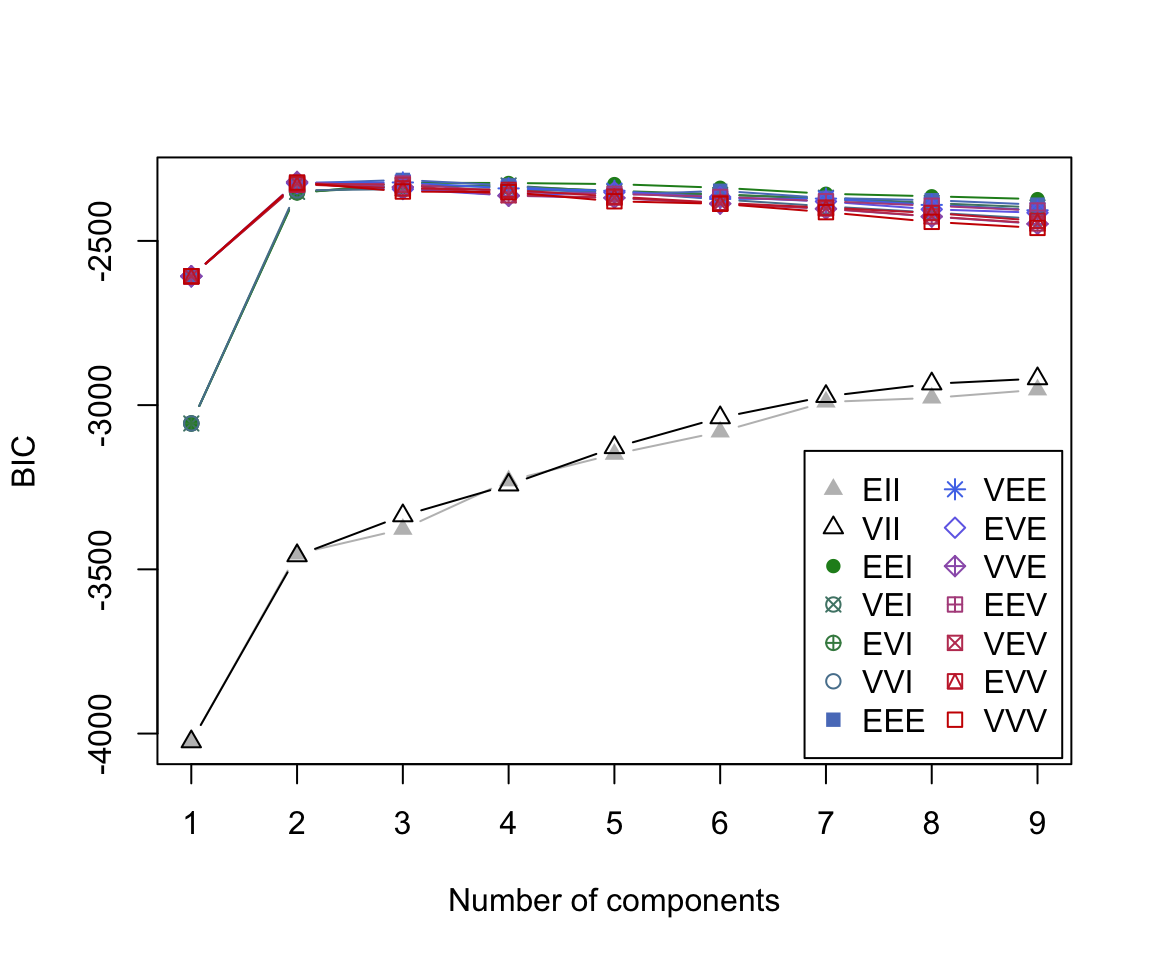
By default, MCLUST will fit a g-component mixture model for all 14 models models for \(g\) = 1, 2, … 9.
For each of these 126 models, a BIC value is calculated.
The BIC
The Bayesian Information Criterion or BIC (Schwarz, 1978) is one of the most widely known and used tools in statistical model selection. The BIC can be written: \[\begin{equation} \text{BIC} = 2 \ell(x, \hat \theta) - p_f \log{n} \end{equation}\] where \(\ell\) is the log-likelihood, \(\hat \theta\) is the MLE of \(\theta\), \(p_f\) is the # of free parameters and \(n\) is the # of observations.
It can be shown that the BIC gives consistent estimates of the number of components in a mixtures model under certain regulator conditions (Keribin, 1988 , 2000)
BIC plot (zoomed)
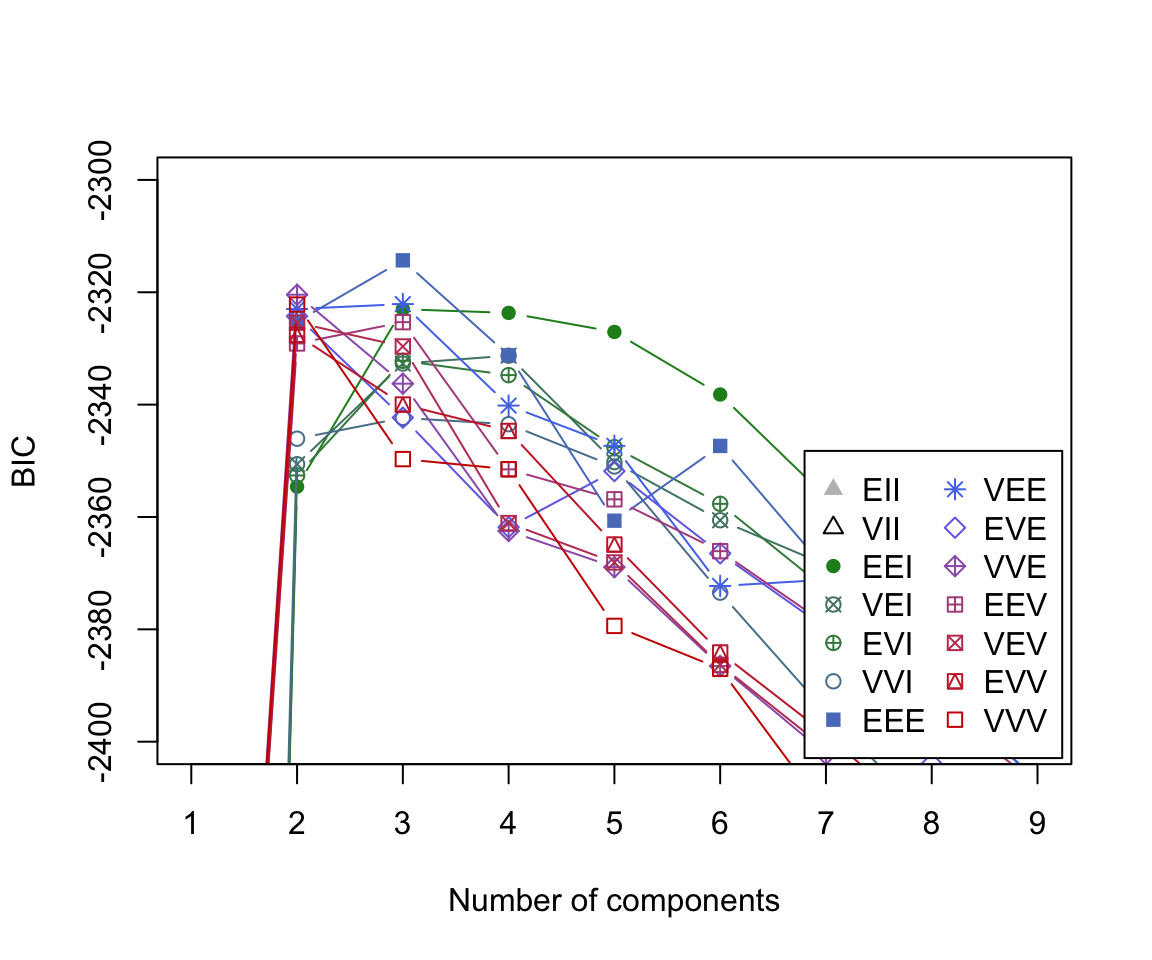
The BIC plot reveals that the model which obtains the largest BIC value is the 3-component mixture model with the ‘EEE’ covariance structure.
Pros
Why Mixture Model-based Clustering?
- Mathematically and statistically sound using standard theory
- Parameter estimation is built-in, along with number of clusters via model selection
- Provides a probabilistic answer to the question “does observation \(i\) belong to group \(G\)?”
- Adaptable, eg. data with outliers/skewness can be handled by non-Gaussian densities
Cons
Cons (a.k.a. open avenues for research)
- Computationally intensive (takes time to estimate and invert covariance matrices; not feasible for small \(n\) large \(p\) without some decomposition)
- Model-selection method is still viewed as an open problem by many practitioners — BIC grudgingly accepted
- Traditional parameter estimation via EM algorithm prone to local maxima, overconfidence in clustering probabilities, and several other issues
Iris

Here i am using “clusters” as groups of data that clump together, but really these are just know classifcation corresponding to the species are Iris setosa, versicolor, and virginica.
jump to the geyser data set.

We can see that these species form distinct clusters (esp in the Sepal.Width vs Petal.Width scatterplot)
Mclust
-
modelNameswhen \(n > d\) all avaible models are fit
The best model (as determined by the BIC) is returned:
modelNameA character string denoting the model at which the optimal BIC occurs.GThe optimal number of mixture components-
parameters:-
profor \(\pi_g\)s,$meanfor \(\mu_g\)s,$variancefor \(\Sigma_g\)s)
-
library(mclust)
# Load example data
data <- iris[, -5]
# Apply Mclust for clustering
model <- Mclust(data)
# Summary of the model
summary(model)----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust VEV (ellipsoidal, equal shape) model with 2 components:
log-likelihood n df BIC ICL
-215.726 150 26 -561.7285 -561.7289
Clustering table:
1 2
50 100