
Data 311: Machine Learning
👻🎃 Lecture 14: Distance Measures 🎃👻
University of British Columbia Okanagan
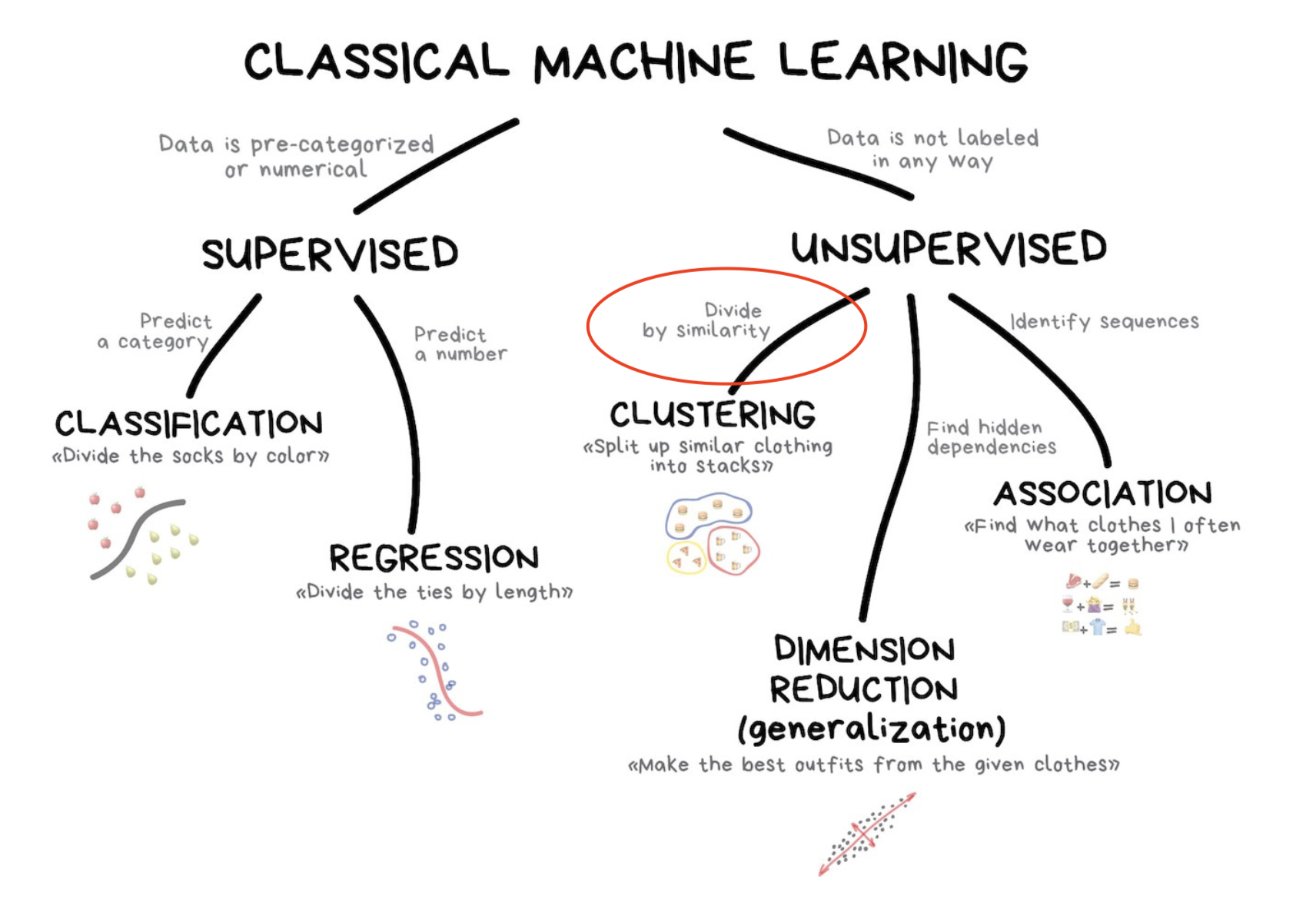
Supervised vs Unsupervised Learning
-
So far we’ve been in the supervised setting where we know the “answers” for response variable \(Y\) which might be continuous (regression) or categorical (classification)
\[ \begin{align} X &\rightarrow Y \\ \quad \text{input data} &\rightarrow \text{output data (``answers")} \end{align} \]
Today we move towards unsupervised learning; that is, problems for which we do not have known “answers”.
Clustering Problem

Potential Solution 1

Potential Solution 2

Potential Solution 3

What is Clustering?
Definition: Clustering is an unsupervised learning technique that groups data points into clusters based on similarity.
Goal: Identify natural groupings or patterns in data without labeled outcomes.
Applications: Market segmentation, image analysis, document organization, and anomaly detection.
Unsupervised complications
- How do we define a cluster?
- How do we determine how many clusters are in the data
- Is there any clusters to be found at all?
- How do we compare two different clustering results?
We’ll tackle some of these problems in upcoming lectures but for now let’s talk about …
Distance
Prior to diving deep on unsupervised methods, it’s important to note that the vast majority are based on distance calculations (of some form).
Distance seems like a straightforward idea, but it is actually quite complex …
For instance, how would the clusetering problem change if we had included some categorical features into the mix?
Outline
Distance Motivation
What is the distance from Trump Tower to Times Square?

What is the distance from Trump Tower to UBCO?
In this case we would need to take into account the curvature of the Earth!
Euclidean Distance
To begin, we’ll consider that all predictors are numeric. The Euclidean distance between two observations \(x_i\) and \(x_j\) is calculated as \[d^{\text{EUC}}_{ij} = \sqrt{\sum_{k=1}^p (x_{ik} - x_{jk})^2}\]
While simple, it is inappropriate in many settings.
Where Euclidean Fails 1
- Consider the following measurements on people:
- Height (in cm),
- Annual salary (in $)
- A $61 difference in annual salary would be considered a minuscule difference, whereas a 61 cm difference in height (approx 2 feet) would be substantial!
Plotted Euclidean Distance

Is it reasonable to assume A is equidistant (at equal distances) with points B and C?
Scale Matters
The scale and range of possible values matters!
If we use a distance-based method, for example KNN, then large scale variables (like salary1) will have a larger effect on the distance between the observations, than variables that are on a small scale (like height).
Hence salary will drive the KNN classification results2.
Standardized Euclidean Distance
One solution is to scale the data to have mean 0, variance 1 via \[z_{ik} = \frac{x_{ik}-\mu_k}{\sigma_k}\] Then we can define standardized pairwise distances as \[d^{\text{STA}}_{ij} = \sqrt{\sum_{k=1}^p (z_{ik} - z_{jk})^2}\]
How it might look after scaling

After scaling those data you might have something more along the lines of this, were C and A are much closer together than A and B
Manhattan Distance
Manhattan distance1 measures pairwise distance as though one could only travel along the axes \[d^{\text{MAN}}_{ij} = \sum_{k=1}^p |x_{ik} - x_{jk}|\]
Similarly, one might want to consider the standardized form \[d^{\text{MANs}}_{ij} = \sum_{k=1}^p |z_{ik} - z_{jk}|\]
Manhattan Visualization

Image sources: Google Maps 2021
Manhattan Visualization 2

The red, yellow, and blue paths all have the same Manhattan distance of 12.
The green line has Euclidean distance of \(6 \sqrt {2} \approx 8.49\) and is the unique shortest path.
Euclidean Visualization

Where Euclidean Fails 2
Euclidean distance can lead to some misleading results when the variables are highly correlated.
Let’s consider data simulated from the bivariate normal distribution having
\[\begin{align} \boldsymbol\mu &= \begin{pmatrix} 2 \\ 3 \end{pmatrix} & \boldsymbol\Sigma &= \begin{pmatrix} 5 & 7\\ 7 & 11 \end{pmatrix} \end{align}\]
Simulation

Which point would you consider more unusual? A or B?
Euclidean Distance for Simulation
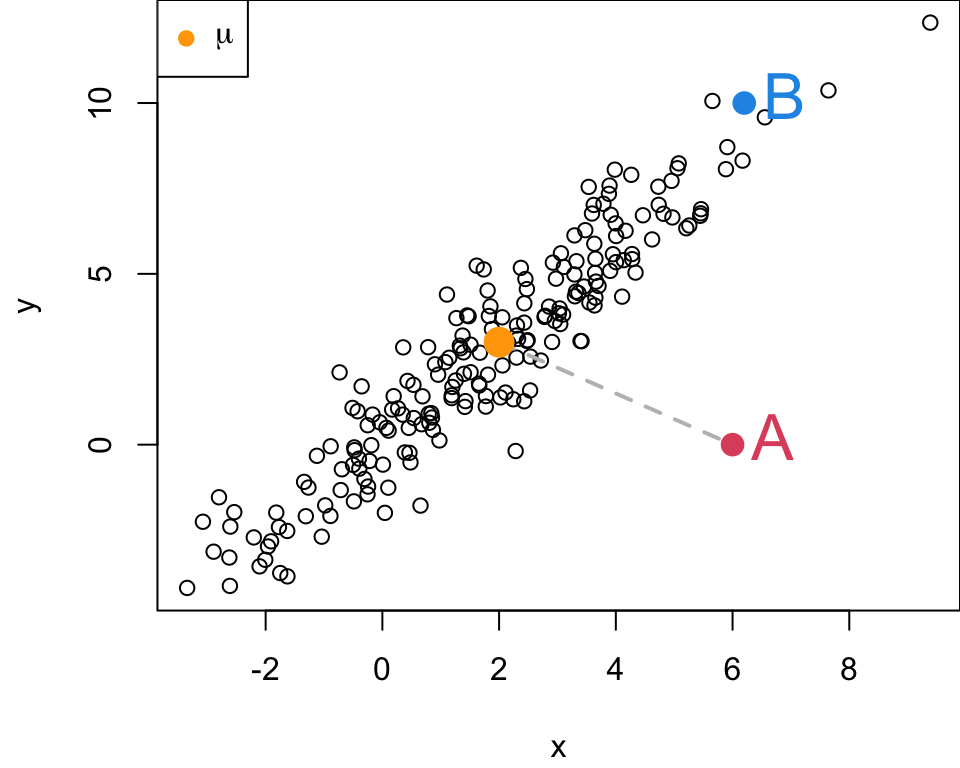
- The euclidean distance between A and \(\mu\) is 5
Euclidean Distance for Simulation

The euclidean distance between A and \(\mu\) is 5
The euclidean distance between B and \(\mu\) is 8.16
B is father in terms of Euclidean distance from the center of this distribution, but it is “closer” to the blob of points than A.
Mahalanobis Distance
Mahalanobis distance takes into account the covariance structure of the data.
It is easiest defined in matrix form \[d^{\text{MAH}}_{ij} = \sqrt{(x_i - x_j)^\top \boldsymbol{\Sigma}^{-1} (x_i - x_j)}\] where \(^\top\) represents the transpose of a matrix and \(^{-1}\) denotes the inverse of matrix.
Mahalanobis Distance for Simulation

- The Mahalanobis distance from point \(\mu\) to A is 64.83
Mahalanobis Distance for Simulation

The Mahalanobis distance from point \(\mu\) to A is 64.83
The Mahalanobis distance from point \(\mu\) to B is 4.57
B is closer to \(\mu\) in terms of Mahalanobis distance than A is, despite A being closer to \(\mu\) in terms of Euclidean distance.
Computing Distances in base R:
Euclidean
[1] 5
[1] 8.163333
A B
B 10.002000
mu 5.000000 8.163333Mahalanobis
Distances with functions
The
dist()function calculates Euclidean, Manhattan, and other standard distances.The
daisy()function in R, from theclusterpackage, calculates dissimilarity matrices for data with mixed types of variables (e.g., numeric, categorical, and ordinal).
dist
1 2 3 4
2 1.7746522
3 0.8753343 2.5110209
4 1.6146343 1.1601774 1.9730709
5 1.2664350 1.8331260 1.2455408 0.8676173 1 2 3 4
2 2.501945
3 1.011478 3.513423
4 1.637059 1.394272 2.648537
5 1.741263 2.082756 1.430667 1.217870Standardization
To standardize our data, we can use the scale() function.
When to Standardize?
- This brings us to an important question…when should we use a standardized measure, and when should we not?
- It is often a good idea, and generally necessary if measurements are on vastly different units.
- Unless you have a good reason to believe that higher variance measures should be weighted heavier, then you probably want to standardize in some form.
iClicker
iClicker: Mahalanobis
Which of the following is a key feature of Mahalanobis distance?
- It considers only the variances of each feature.
- It can handle both numeric and categorical features.
- It only works with binary features.
- It accounts for both variances and covariances between features.
From Numeric to Mixed
What if some/all the predictors are not numeric?
We can consider several methods for calculating distances based on matching.
For binary data case, we can get a “match” either with a 1-1 agreement or 0-0.
Matching Binary Distance
- We can actually define another matching method that is equivalent to Manhattan/city-block,
\[d^{\text{MAT}}_{ij} = \text{\# of variables with opposite value}\]
This is simple sum of disagreements is the unstandardized version of the M-coefficient.
Let’s see an example,…
President Example
Let’s look at some binary variables for US presidents:
-
Democratlogical indicating if they are a democrat -
Govenorlogical indicating if they were they formerly a governor 1 = yes -
VPlogical indicating if they were formerly a vice president -
2nd Termlogical indicating if they serve a second term -
From Iowalogical indicating if they are originally from Iowa
Distance between Bush and Obama
| Democrat | Governor | VP | 2nd Term | From Iowa | |
|---|---|---|---|---|---|
| GWBush | 0 | 1 | 0 | 1 | 0 |
| Obama | 1 | 0 | 0 | 1 | 0 |
| Trump | 0 | 0 | 0 | 0 | 0 |
The Manhattan distance between GWBush vs Obama: \[|0-1|+|1-0|+|0-0|+|1-1|+|0-0| = 2\]
This is equivalent to the Matching Binary Distance since they do not match in two variables: Democrat and Governor. \[
d^{\text{MAT}}_{12} = \text{\# of variables with opposite value} = 2
\]
M-coefficient
The M-coefficient (or matching coefficient) is simply the proportion of variables in disagreement in two objects over the total number of variable comparisons, p.
To put another way, you can think of Matching Binary Distance as an unscaled version of the M-coefficient.
\[ \begin{equation} d_{12}^{\text{M-coef}} = \frac{d_{12}^{MAT}}{p} = \frac{2}{5} = 0.4 \end{equation} \]
Calculate M-coefficient
| Democrat | Governor | VP | 2nd Term | From Iowa | |
|---|---|---|---|---|---|
| GWBush | 0 | 1 | 0 | 1 | 0 |
| Obama | 1 | 0 | 0 | 1 | 0 |
| Trump | 0 | 0 | 0 | 0 | 0 |
Calculating the simple matching M-coefficient1 \[d^{\text{M-coef}}_{12} = \frac{\text{\# of disagreements}}{p} = \frac{2}{5} = 0.4\]
Question: Do you think a 0-0 match in
From Iowashould really map to a binary distance of 0?
| Democrat | Governor | VP | 2nd Term | From Iowa | |
|---|---|---|---|---|---|
| GWBush | 0 | 1 | 0 | 1 | 0 |
| Obama | 1 | 0 | 0 | 1 | 0 |
| Trump | 0 | 0 | 0 | 0 | 0 |
Since a 0-0 “match” does not imply that they are from similar places in the US, I would argue not.
Asymmetric Binary Distance
For reasons discussed on the previous slide, it makes sense to toss out those 0-0 matches.
In this case, what we would be considering is asymmetric binary distance
\[d^{\text{ASY}}_{ij} = \frac{\text{\# of 0-1 or 1-0 pairs}}{\text{\# of 0-1, 1-0, or 1-1 pairs}}\]
Calculate Asymmetric Distance
| Democrat | Governor | VP | 2nd Term | From Iowa | |
|---|---|---|---|---|---|
| GWBush | 0 | 1 | 0 | 1 | 0 |
| Obama | 1 | 0 | 0 | 1 | 0 |
| Trump | 0 | 0 | 0 | 0 | 0 |
The asymmetric binary distance between Bush and Obama:
\[ \begin{align} d^{\text{ASY}}_{ij} &= \frac{\text{\# of 0-1 or 1-0 pairs}}{\text{\# of 0-1, 1-0, or 1-1 pairs}}= \frac{2}{3} = 0.67 \end{align} \]
iClicker
Asymmetric Binary Distance
Which of the following best describes the Asymmetric Binary Distance
- It ignores 0-0 matches when calculating the distance between two binary vectors.
- It calculates the number of matching variables divided by the total number of variables.
- It only considers Euclidean distances on standardized binary data.
- It includes 0-0 matches to fully count the differences between two binary vectors.
Distance for Qualitative Variables
For categorical variables having \(>2\) levels, a common measure is essentially standardized matching once again.
If \(u\) is the number of variables that match between observations \(i\) and \(j\) and \(p\) is the total number of variables, the measure1 is calculated as: \[d^{\text{CAT}}_{ij} = 1 - \frac{u}{p}\]
Distance for Mixed Variables
But often data has a mix of variable types.
Gower’s distance is a common choice for computing pairwise distances in this case.
The basic idea is to standardize each variable’s contribution to the distance between 0 and 1, and then sum them up.
Gower’s Distance
Gower’s distance (or Gower’s dissimilarity) is calculated as: \[d^{\text{GOW}}_{ij} = \frac{\sum_{k=1}^p \delta_{ijk} d_{ijk}}{\sum_{k=1}^p \delta_{ijk}}\]
where
-
\(\delta_{ijk}\) = 1 if both \(x_{ik}\) and \(x_{jk}\) are non-missing1 (0 otherwise),
- \(d_{ijk}\) depends on variable type
Quantitative numeric \[d_{ijk} = \frac{|x_{ik} - x_{jk}|}{\text{range of variable k}}\]
Qualitative (nominal, categorical) \[d_{ijk} = \begin{cases} 0 &\text{ if obs $i$ and $j$ agree on variable $k$, } \\ 1 &\text{ otherwise} \end{cases} \]
Binary \(d_{ijk} = \begin{cases} 0 & \text{ if obs $i$ and $j$ are a 1-1 match, } \\ 1 &\text{ otherwise} \end{cases}\)
Example of Gower’s Distance
| Party | Height (cm) | Eye Colour | 2 Terms | From Iowa | |
|---|---|---|---|---|---|
| Biden | Democratic | 182 | Blue | No | No |
| Trump | Republican | 188 | Blue | NA | No |
| Obama | Democratic | 185 | Brown | Yes | No |
| Fillmore | Third Party | 175 | Blue | No | No |
Distance between Biden and Trump
0-0 pairs not counted as matches
We will not consider 0-0 to be matches.
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | ||
Height |
1 | ||
EyeColour |
1 | ||
2 Terms |
0 | ||
Iowa |
0 |
Party Calculations
| Party | Height (cm) | Eye Colour | 2 Terms | From Iowa | |
|---|---|---|---|---|---|
| Biden | Democratic | 182 | Blue | No | No |
| Trump | Republican | 188 | Blue | NA | No |
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | 1 | 1 |
Height |
1 | ||
EyeColour |
1 | ||
2 Terms |
0 | ||
Iowa |
0 |
Range of Presidents Heights
To calculate \(d_{12k}\) we need the range of the Height(cm)s
- Tallest President: Abraham Lincoln at 193 centimeters
- Shortest President: James Madison at 163 centimeters
Range of Height variable (\(k = 2\)) is 193-163 = 30
\(d_{1,2,2} = \dfrac{|x_{ik} - x_{jk}|}{\text{range of variable k}} = \dfrac{|188 - 182|}{30} = 0.2\)
Height Calculations
| Party | Height (cm) | Eye Colour | 2 Terms | From Iowa | |
|---|---|---|---|---|---|
| Biden | Democratic | 182 | Blue | No | No |
| Trump | Republican | 188 | Blue | NA | No |
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | 1 | 1 |
Height |
1 | 0.2 | 0.2 |
EyeColour |
1 | ||
2 Terms |
0 | ||
Iowa |
0 |
EyeColour Calculations
| Party | Height (cm) | Eye Colour | 2 Terms | From Iowa | |
|---|---|---|---|---|---|
| Biden | Democratic | 182 | Blue | No | No |
| Trump | Republican | 188 | Blue | NA | No |
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | 1 | 1 |
Height |
1 | 0.2 | 0.2 |
EyeColour |
1 | 0 | 0 |
2 Terms |
0 | ||
Iowa |
0 |
2 Terms Calculations
| Party | Height (cm) | Eye Colour | 2 Terms | From Iowa | |
|---|---|---|---|---|---|
| Biden | Democratic | 182 | Blue | No | No |
| Trump | Republican | 188 | Blue | NA | No |
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | 1 | 1 |
Height |
1 | 0.2 | 0.2 |
EyeColour |
1 | 0 | 0 |
2 Terms |
0 | - | 0 |
Iowa |
0 |
From Iowa Calculations
| Party | Height (cm) | Eye Colour | 2 Terms | From Iowa | |
|---|---|---|---|---|---|
| Biden | Democratic | 182 | Blue | No | No |
| Trump | Republican | 188 | Blue | NA | No |
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | 1 | 1 |
Height |
1 | 0.2 | 0.2 |
EyeColour |
1 | 0 | 0 |
2 Terms |
0 | - | 0 |
Iowa |
0 | 0 | 0 |
Gower’s Distance Calculation
| \(k\) | \(\delta_{12k}\) | \(d_{12k}\) | \(\delta_{12k}d_{12k}\) |
|---|---|---|---|
Party |
1 | 1 | 1 |
Height |
1 | 0.2 | 0.2 |
EyeColour |
1 | 0 | 0 |
2 Terms |
0 | - | 0 |
Iowa |
0 | 0 | 0 |
| Total | 3 | 1.2 |
\[ d^{\text{GOW}}_{ij} = \frac{\sum_{k=1}^p \delta_{ijk} d_{ijk}}{\sum_{k=1}^p \delta_{ijk}} = \frac{1.2}{3} = 0.4 \]
Comments
Gower distance will always be between 0.0 and 1.0
a distance of 0.0 means the two observations are identical for all non-missing predictors
a distance of 1.0 means the two observations are as far apart as possible for that data set
The Gower distance can be used with purely numeric or purely non-numeric data, but for such scenarios there are better distance metrics available.
There are several variations of the Gower distance, so if you encounter it, you should read the documentation carefully.