Data 311: Machine Learning
Lecture 11: Bootstrap Aggregating (Bagging) and Random Forests
University of British Columbia Okanagan
Introduction
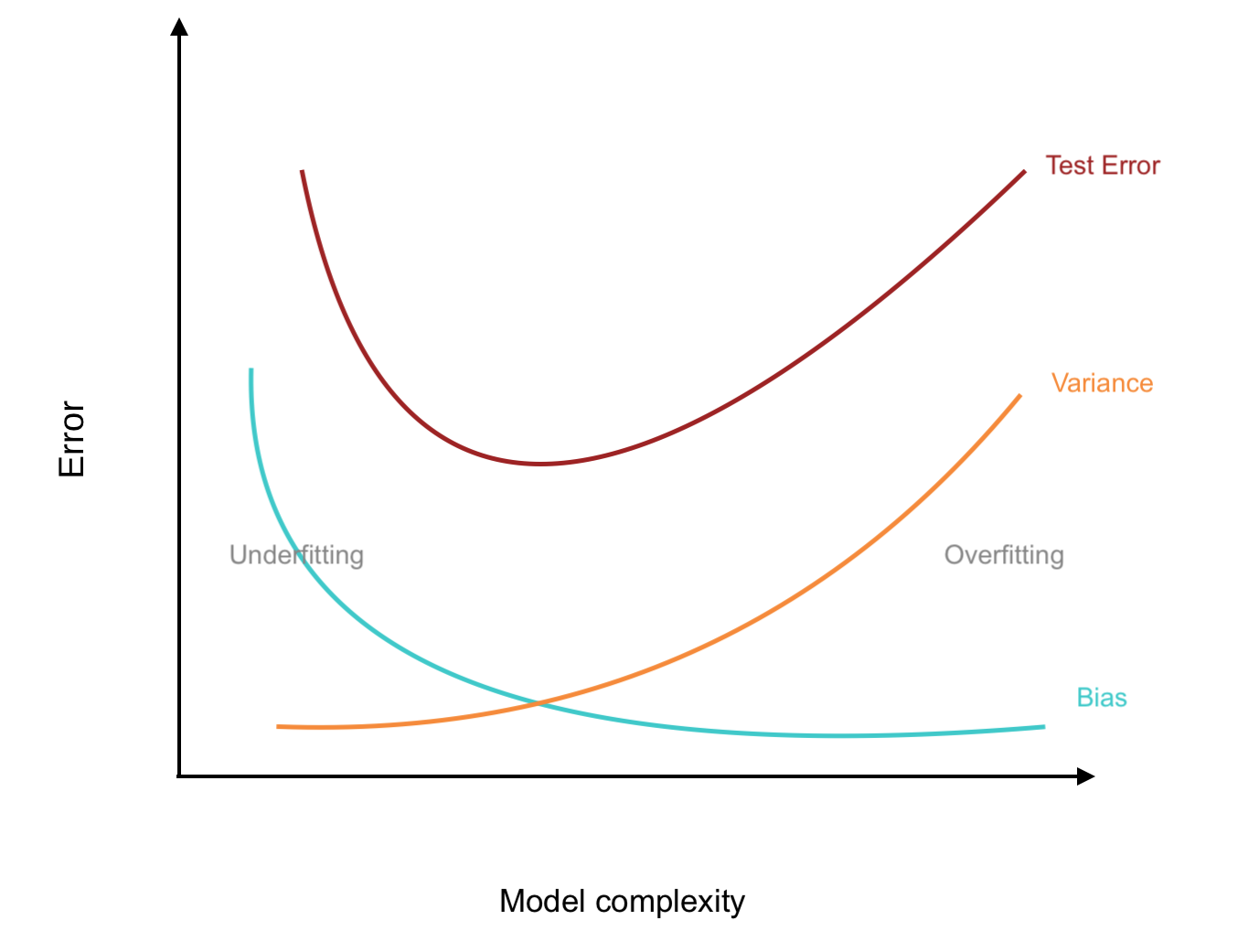
Trees have one aspect that prevents them from being ideal tool for predictive learning, namely inaccuracy. - ESL
We’ve just wrapped up some discussion on decisions trees, or CARTs which suggests that they are not particularly “good” models in many scenarios.
Why? Bushy trees tend to have high variance and pruned trees have high bias
Preface
Today we’ll be looking at some ensemble methods to make them more competitive:
Namely, we will be looking at two embsemble methods:
Bagging, which combines multiple models trained on random subsets to reduce variance
Boosting which sequentially builds models that correct previous errors to reduce bias
Preface
Namely, we will be looking at two embsemble methods:
-
Bagging, which combines multiple models trained on random subsets to reduce variance
- We’ll also look at Random forests, a specific type of bagging technique with an additional layer of randomness to improve model performance.
Boosting which sequentially builds models that correct previous errors to reduce bias (covered next lecture)
Bushy Trees

Goal of Bagging


Stumps


Goal of Boosting


Ensemble Methods
Ensemble methods are techniques that combine the predictions of multiple models to improve overall predictive performance. Then can be divided in two groups:
-
Parallel Learners: multiple models are trained independently of each other.
- e.g. Bootstrap Aggregation (Bagging), Random Forests
-
Sequential learners: models are trained sequentially, one after another. Each model in the sequence is typically trained to correct the errors made by the previous models.
- e.g. Boosting, AdaBoost, Gradient Boosting, XGBoost
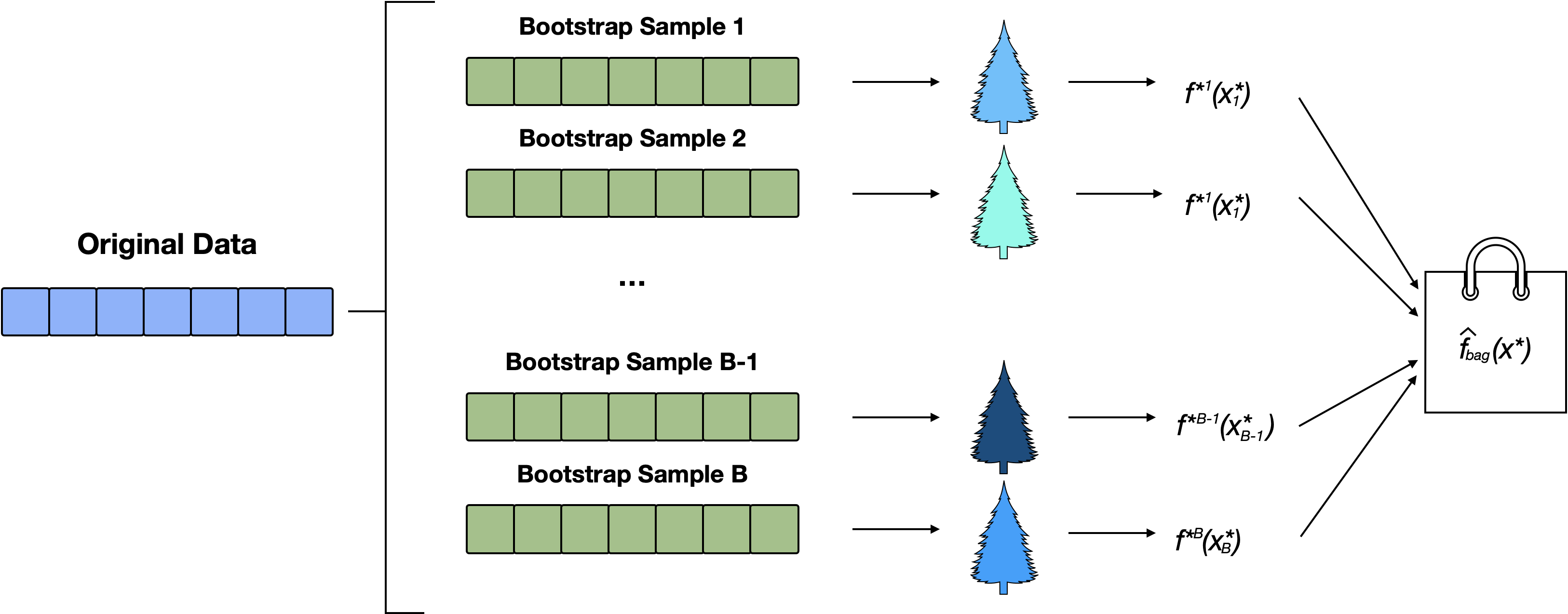
Bagging
Bagging (Bootstrap Aggregating) combines multiple models trained on different bootstrap samples (random subsets with replacement) to reduce variance.
Because CART models tend to overfit the training data, we will use bagging to combine multiple “bushy” trees.
This approach creates a more robust and generalizable model with lower variance than any single CART model.
General bagging
Generating \(B\) bootstrapped samples
Train \(B\) models, \(\hat f^{*b}\) for \(b = 1, \dots B\),
Aggregate the predictions
For regression this is simply the average prediction:
\[ \begin{equation} \hat f_{bag}(x) = \frac{\sum_{b=1}^B f^{*b}(x^*_b)}{B} \end{equation} \]

The main idea is to pool a collection of random decision trees

Bagging Schematic
Model Assessment
We could get an estimate of error using our test set, however, we can get a different estimate “for free”:
Recall: a bootstrap sample consists of sampling \(n\) observations with replacement from our training set1
As we are sampling with replacement, some observations are randomly left out while others are duplicated.
It can be shown that for each bootstrap sample, on average roughly \(\frac{1}{3}\) of the observations will not appear …
Probability calculations
\[ \Pr(x_i \text{ is not selected in the bootstrap sample}) \]
\[ \begin{align} = P(x_i & \text{ is not selected on first draw}) \times \dots \\ & \quad \dots \times P(x_i \text{ is not selected on $n^{th}$ draw}) \end{align} \]
\[ \begin{align} &= \left(\frac{n-1}{n}\right)\times \dots \times \left(\frac{n-1}{n}\right) = \left(1 - \frac{1}{n}\right)^n \\ \end{align} \]
Now if we take the limit as \(n \to \infty\) we get:
\[ \begin{equation} \lim_{n \to \infty} \left(1 - \frac{1}{n}\right)^n = \frac{1}{e} \approx \frac{1}{2.7183} \approx \frac{1}{3} \end{equation} \] where \(e\) is the exponential constant approximately 2.718
Bootstrap Visualization
Image source: rasbt.github

The figure above illustrates how three random bootstrap samples drawn from an ten-sample dataset \((x_1,x_2,...,x_{10})\) and their ‘out-of-bag’ sample for testing may look like.
OOB observations
The observations not included in the bootstrap sample during bagging are called the Out of Bag (OOB) observations.
These OOB observations can serve a similar role as the validation sets in cross-validation
By serving as internal validation, OOB provides an efficient way to evaluate model performance, making it a practical alternative to cross-validation.
Recall 5-fold CV Visualization

Aggregating OOB Predictions
For each training observation \(x_i\), predictions are made using only the models where \(x_i\) was an OOB observation1.
-
For regression, the OOB prediction for \(x_i\) is the average predictions from the OOB models:
\[ \hat{y}_i^{\text{OOB}} = \frac{1}{B_i} \sum_{b \in \text{OOB}i} \hat{y}_{i}^{(b)} \]
For classification, the OOB prediction is determined by majority voting among the models for which \(x_i\) was OOB.
OOB Error Estimation
For regression we calculate the Mean Squared Error (MSE): \[ \text{OOB MSE} = \frac{1}{n} \sum_{i=1}^{n} (y_i - \hat{y}_i^{^\text{OOB}})^2 \] where \(y_i\) is the true value and \(\hat{y}_i^{\text{OOB}}\) is the OOB prediction for observation \(x_i\). For classification we calculate the misclassification rate: \[ \text{OOB Error} = \frac{1}{n} \sum_{i=1}^{n} I(y_i \neq \hat{y}_i^{\text{OOB}}) \]
So if we wanted to predict the response for \(x_3\) we would use the trees fitted to Bootstrap sample 1 and Bootstrap sample 3 and average the result.

Key Points:
- OOB observations are those that are not included in the bootstrap sample used for training each model.
- OOB error provides an internal validation estimate of the test error, similar to cross-validation but without explicitly creating validation sets.
- The OOB estimate is efficient and a reliable alternative to cross-validation, particularly for large datasets.
iClicker
Number of Bootstrap observations
On average, what proportion of the training dataset is included in each bootstrap sample used to create the training set in bagging?
roughly 1/3
roughly 2/3 ✅ Correct
roughly \(n\), the number of samples in the training set
exactly, \(n\), the number of samples in the training set
iClicker
Purpose of bagging
What is the main purpose of bagging in ensemble methods?
To increase bias
To reduce variance ✅ Correct
To increase overfitting
To improve interpretability
iClicker
OOB observations
What is the main purpose of using the bootstrap in the context of bagging?
To create varied samples of the training data ✅ Correct
For estimating the test error without needing a separate validation set
To make the model deterministic
For feature selection
Example: Beer
Recall the beer data from last lecture (names removed):
69 observations and 7 columns
An example of six decision trees independently trained on bootstrap samples of the beer dataset. Each tree is slightly different due to the unique bootstrap sample used for training. This variability helps reduce model variance when predictions are aggregated across all (e.g. 500) trees. Compare with random forests
Example: Beer OLS Regression
OLS Regression:
Regression tree

Cross-validation Errors
Let’s calculate the CV RSS error for OLS Regression:
# Get the CV estimate of MSE (two ways)
rss_lm = sum((beercv$price-beercv$cvpred)^2) # 10-fold CV RSS for lm
cv_mse <- attr(beercv, "ms")
cv_mse*nrow(beerdat) # same as rss_lm above[1] 57.33652and the (actual) CV RSS error estimate for Regression Trees…
Bagging Regression Trees in R
library(randomForest); set.seed(134891)
p = 5 # number of predictors to consider at each split
(beerbag <- randomForest(price~., data=beerdat, ntree=500, mtry = p)) ...
Type of random forest: regression
Number of trees: 500
No. of variables tried at each split: 5
Mean of squared residuals: 0.9338468
% Var explained: 54.71
...Comparison
Model price~.
|
Estimate of Error (RSS) |
|---|---|
| Linear model | 57.34 (from CV) |
| (Single) Regression Tree | 77.57 (from CV) |
| (500) Bagged Regression Trees | 64.44 (from OOB) |
Error Comments
It’s worth noting that comparison we just made…
Technically, we have not performed cross validation on our bagged trees, and yet we are comparing the (OOB) error to properly cross-validated errors for LM and CART
OOB estimation is a similar (and more efficient) way for providing realistic long term error estimates
Takeaway: For practical purposes, OOB error from bagged models can be compared to CV error from other models.
Variable Importance
- The ensemble of trees helps identify which variables are most influential in the model.
- Another thing we get “for free” in bagging decision tress is a measure of variable importance.
- To quantify variable importance, we measure how much each variable improves the model’s predictions.
Variable Importance in R
- We can obtain an overall summary of the importance of each predictor if requested using:
Two measures are provided for variable “importance”1.
The plot and values are produced using the following:
Bagging Variable Importance Plots

Jump to RF varImpPlot
%IncMSE
Calculate the OOB MSE for the full set of \(B\) trees in your random forest, say, \(\text{MSE}_{full}\)
For the variable in question, randomly shuffle the observations (leave the order correct for the other columns).
Carry out predictions, calculate \(\text{MSE}_{shuff}\) then check the change from the full version for the variable in question: \[\begin{align} \text{%IncMSE} = \frac{\text{MSE}_{{shuff}}-\text{MSE}_{full}}{\text{MSE}_{full}} \end{align}\]
IncNodePurity
IncNodePurityis the total increase in node purity obtained from splitting on the variable, averaged over all trees.In other words,
IncNodePurityis the increase in homogeneity in the data partitions.For regression, “purity” is measured by residual sum of squares (RSS)1.
A higher increase of node purity indicates that the variable is more important.
Bagging vs Single Tree
Cons
❌ We no longer have one model, but an average of many models!
❌ Bagging looses the interpretability of our model
❌ It is comparatively slower than fitting a single tree
Pros
✅ Averaging prevents overfiting
✅ improves prediction
✅ OOB error estimate “for free”
✅ Variable importance “for free”
✅ Doesn’t require CV/test set
Random forests
Random Forests (RF) improve upon bagging by introducing a small tweak that decorrelates the trees.
Like bagging, RF build a number of trees on bootstrapped training samples; however, each time a split in a tree is considered, a random sample of \(m\) predictors (where \(m < p\) = the total number of predictors) is considered.
The default for classification is \(m = \sqrt{p}\) where \(p\) is number of predictors, and for regression \(m = p/3\)
RF Schematic
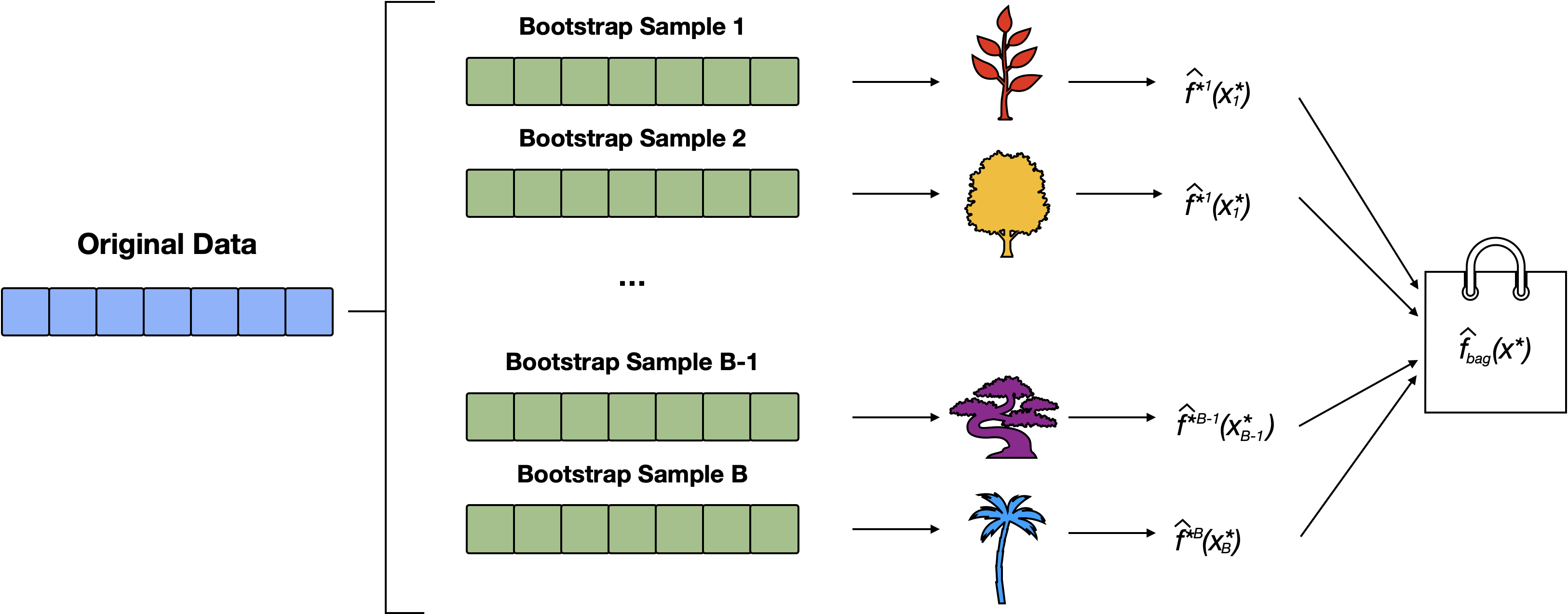
RF build on bagging by training multiple trees on different bootstrap samples and adding an extra layer of randomness by selecting a random subset of features at each split. This approach results in more diverse trees and better model generalization. Compare with Bagging Schematic
Motivating example
Suppose that there is one very strong predictor in the data set (eg.
bitterin thebeerexample)In the collection of bagged trees, most or all of the trees will likely use this strong predictor in the top split, hence, all of the bagged trees will look quite similar to each other.
As discussed in our CV lecture, averaging highly correlated models does not lead to as large of a reduction in variance as averaging many uncorrelated quantities.

An example of six decision trees independently trained on bootstrap samples of the beer dataset in a RF. Each tree is different due to the unique bootstrap sample and the random selection of features at each split. This variability helps reduce model variance when predictions are aggregated across all (e.g., 500) trees. Compare with bagging
Steps for RF
Create \(B\) bootstrap samples
-
Fit a tree* to each bootstrap sample,
* when creating nodes in each decision tree, only a random subset of \(m\) features is considered for splitting at each node.
Each decision tree is used to make a prediction.
Aggregate the \(B\) predictions.
R function
We use the randomForest() function from the randomForest package to implement bagging and RF. Usage:
-
ntree: the number of trees to grow (default = 500) -
mtry: the number of variables randomly selected at each node (default in classification is \(\sqrt{p}\) and \(p/3\) in regression; where \(p\) is the number of predictors). -
importanceShould importance of predictors be assessed?
Optimizing the RF
How do we determine how many subsets of features to consider at each split?
mtryis a hyperparameter of our model and it’s choice can impact the performance of our random forest.In practice the default values are reasonable starting point and often works well in practice.
Random Forest on beer
mtry = \(\sqrt{p}\) by default1
...
Type of random forest: regression
Number of trees: 500
No. of variables tried at each split: 2
Mean of squared residuals: 0.8469584
% Var explained: 58.93
...The OOB estimate for RSS = MSE\(\times n\) = 0.847 * 69 = 58.44
Comparison
| Method | RSS Estimate |
|---|---|
| Linear model | 57.34 |
| Random Forest | 58.44 |
| Bagging | 64.44 |
| Single Tree | 77.57 |
For the beer data set, RF does better than single tree and bagging; however, it does still not outperform our linear regression model.
RF Variable Importance Plot

Jump to Bagging varImpPlot
iClicker
RF vs bagging
In Random Forests, what additional step is taken compared to bagging to further reduce overfitting?
Training on larger dataset
Averaging the results of multiple models
Randomly selecting a subset of features at each split ✅ Correct
Using a single tree for predictions
iClicker
OOB error
What does the term “Out-of-Bag (OOB)” error refer to in the context of Random Forests and Bagging?
Error on a separate test dataset
Error on the original training set
Error calculated on the samples not used in the bootstrap sample ✅ Correct
Error on a validation dataset
Classification Example
The smaller (more common) Italian red
winedata set178 samples of wine from three varietals (Barolo, Barbera, Grignolino) grown in the Piedmont region of Italy with 13 chemical measurements, i.e. \(p=13\)
Let’s compare a single classification tree to a tree found using bagging and random forests …



Single Decision Tree

Cost-complexity pruning

Pruned Classification Tree

Cross-validation error estimate
Let’s obtain the test error (in this case misclassification rate) by cross-validation at the minimum value of \(\alpha\):
Single pruned tree results

Bagging wine
Here we are saying we want to do bagging
mtry= \(m = p\)The
importanceargument tells the aglorithm that we want it to compute variable importance.
Note that now, we don’t have a single tree which we can plot.
Bagging Results
Call:
randomForest(formula = factor(Class) ~ ., data = wine, mtry = 13, importance = TRUE)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 13
OOB estimate of error rate: 3.37%
Confusion matrix:
1 2 3 class.error
1 57 2 0 0.03389831
2 1 68 2 0.04225352
3 0 1 47 0.02083333The OOB error rate of 3.37% versus 8.427% for a standard tree
Bagging varImpPlot wine

Jump to RF varImpPlot
Random Forests
# use default: mtry = floor(sqrt(p)) = floor(sqrt(13)) = floor(3.605551)
(wineRF <- randomForest(factor(Class)~., # ntree = 500,
data=wine, importance=TRUE))...
Number of trees: 500
No. of variables tried at each split: 3
OOB estimate of error rate: 1.12%
Confusion matrix:
1 2 3 class.error
1 59 0 0 0.00000000
2 1 69 1 0.02816901
3 0 0 48 0.00000000
...\(\quad\) Simple Tree (8.43%) < Bagging (3.37%) < RF (1.12%)
RF varImpPlot wine

Jump to bagging varImpPlot
Summary of RF
Random forests share many of the same pros and cons as Bagging vs Single Tree.
They are considered one of the best “off-the-shelf” machine learning models due to their strong predictive performance.
Random forests require minimal hyperparameter tuning (mainly the number of features to consider at each split,
mtryor \(m\)), and the default settings usually perform well in most cases.
Comments
These trees for each bootstrap sample are grown deep (i.e. “bushy”), and are not pruned.
Consequently, each individual tree will have high variance, but low bias
The textbook suggests \(B=100\) as generally sufficient, but probably 500 to 1000 is safer (500 is the default).