Data 311: Machine Learning
Lecture 9: Bootstrap
University of British Columbia Okanagan
Introduction
Last lecture we saw a resampling method called cross-validation (CV) for estimating test error.
Now, a resampling method called the bootstrap where we repeatedly sample from a dataset with replacement.
This allows us to estimate the distribution of a statistic (such as a standard deviation) when we only have limited data.
Hence, it can quantify the uncertainty associated with a given estimator1 or statistical learning method.
Motivation
Sometimes it is possible to work out things like bias and standard error (SE) of an estimator in closed form.
For example the standard error of \(\overline X\), the estimator for the mean of a normally distributed population with having (unknown population) mean \(\mu\) and variance \(\sigma\), is \[\text{SE}(\overline X) = \frac{\sigma}{\sqrt{n}}\]
Sampling Distribution of \(\overline X\)
More specifically if \(X_i\) are i.i.d1 Normal(\(\mu\), \(\sigma^2\)), then \(\overline X=\frac{1}{n}\sum_{i=1}^n X_i\) has the sampling distribution given by:
\[ \overline X \sim \text{Normal}(\mu, \frac{\sigma}{\sqrt{n}}) \]
Recall that the standard error of an estimator simply refers to the standard deviation of the sampling distribution of that estimator.
Motivation (cont’d)
From this information we could create 95% confidence intervals for our estimate of \(\mu\)
\[ \begin{equation} \bar{x}\ \pm \ t_{0.025, n-1} \left(s/\sqrt{n}\right) \end{equation} \]
- \(\bar{x}\) is the sample mean.
- \(t_{\frac{\alpha}{2}, \, n-1}\) is the critical value from the \(t\)-distribution with \(n-1\) degrees of freedom
- \(s\) is the sample standard deviation.
- \(n\) is the sample size.
Sampling Distribution
Generally speaking a sampling distribution is the probability distribution of a statistic (or estimator) that is obtained through repeated sampling of a specific population.
In simulations, we could generate large number of samples and record the value of the statistic of interest to obtain an empirical estimate of the sampling distribution.
In many contexts, only one sample is observed, but the sampling distribution can be found theoretically.
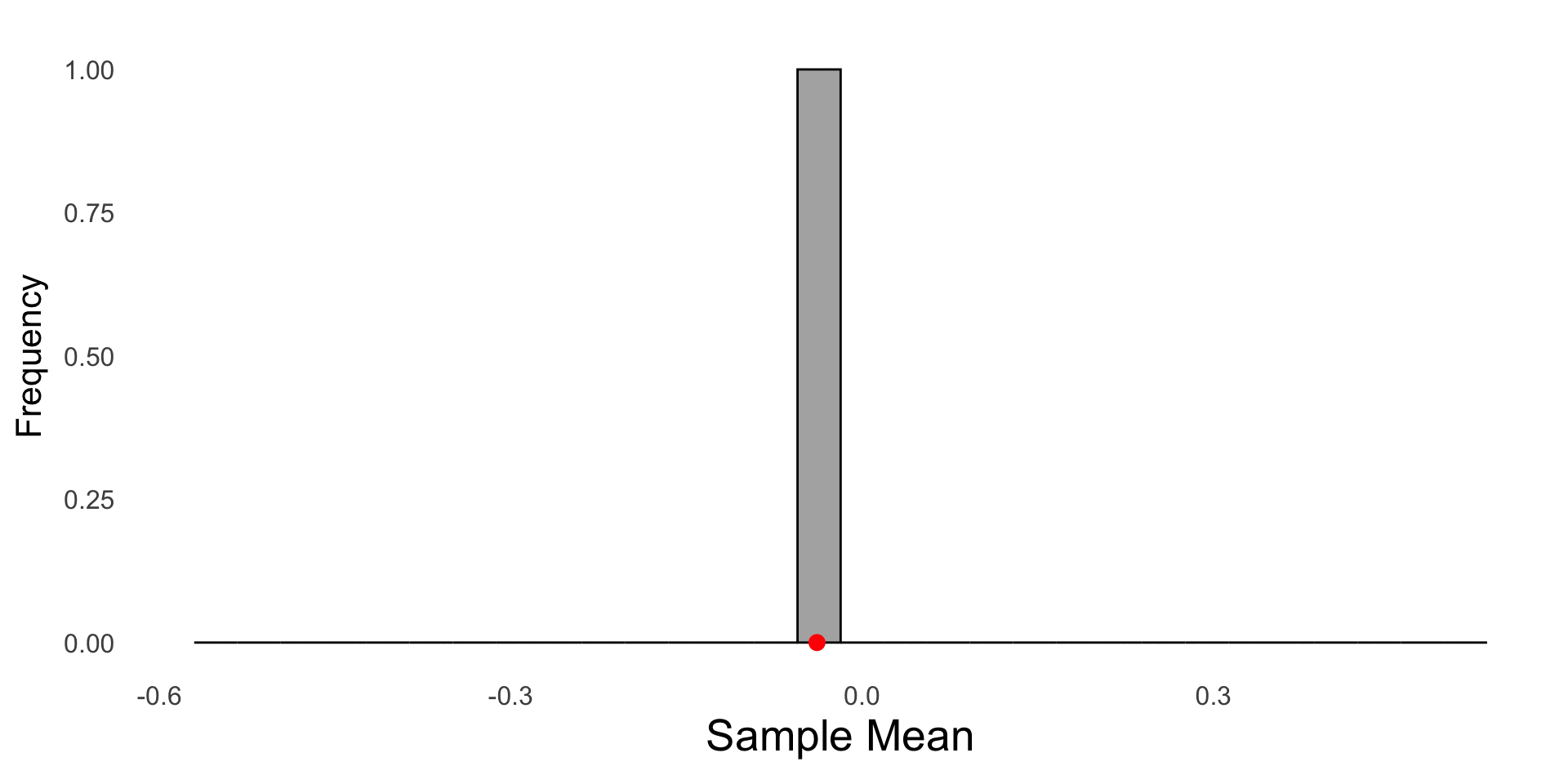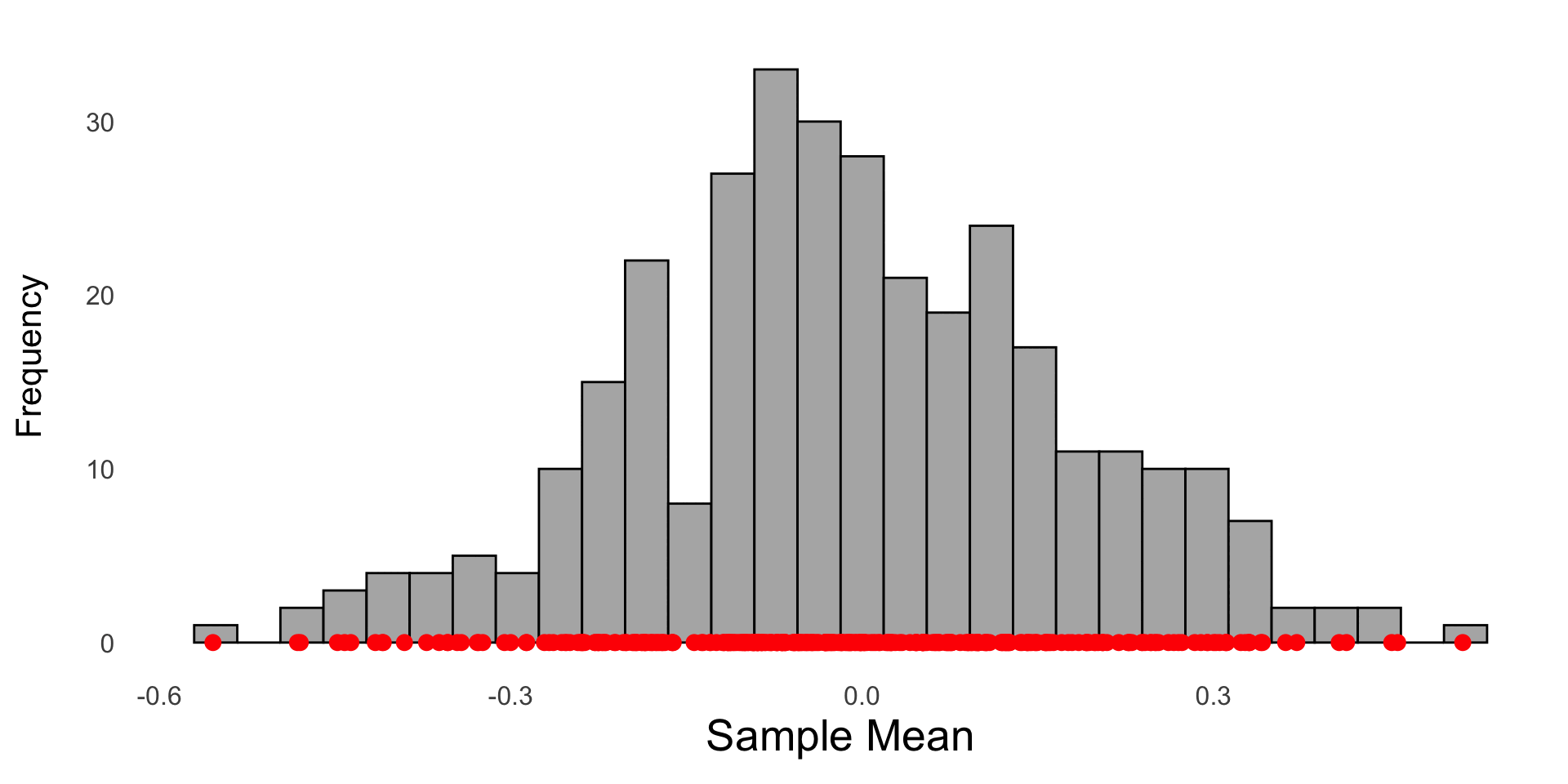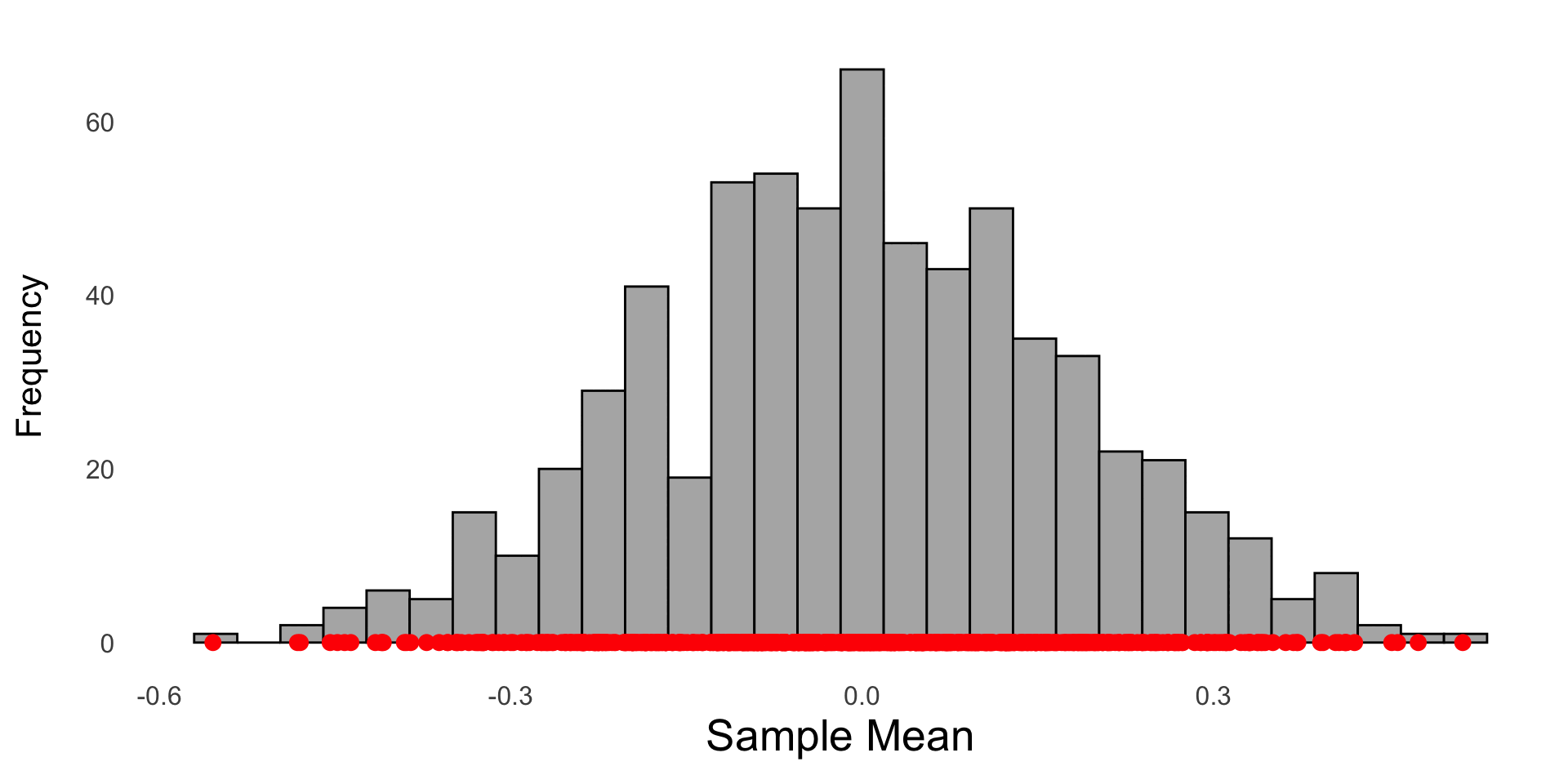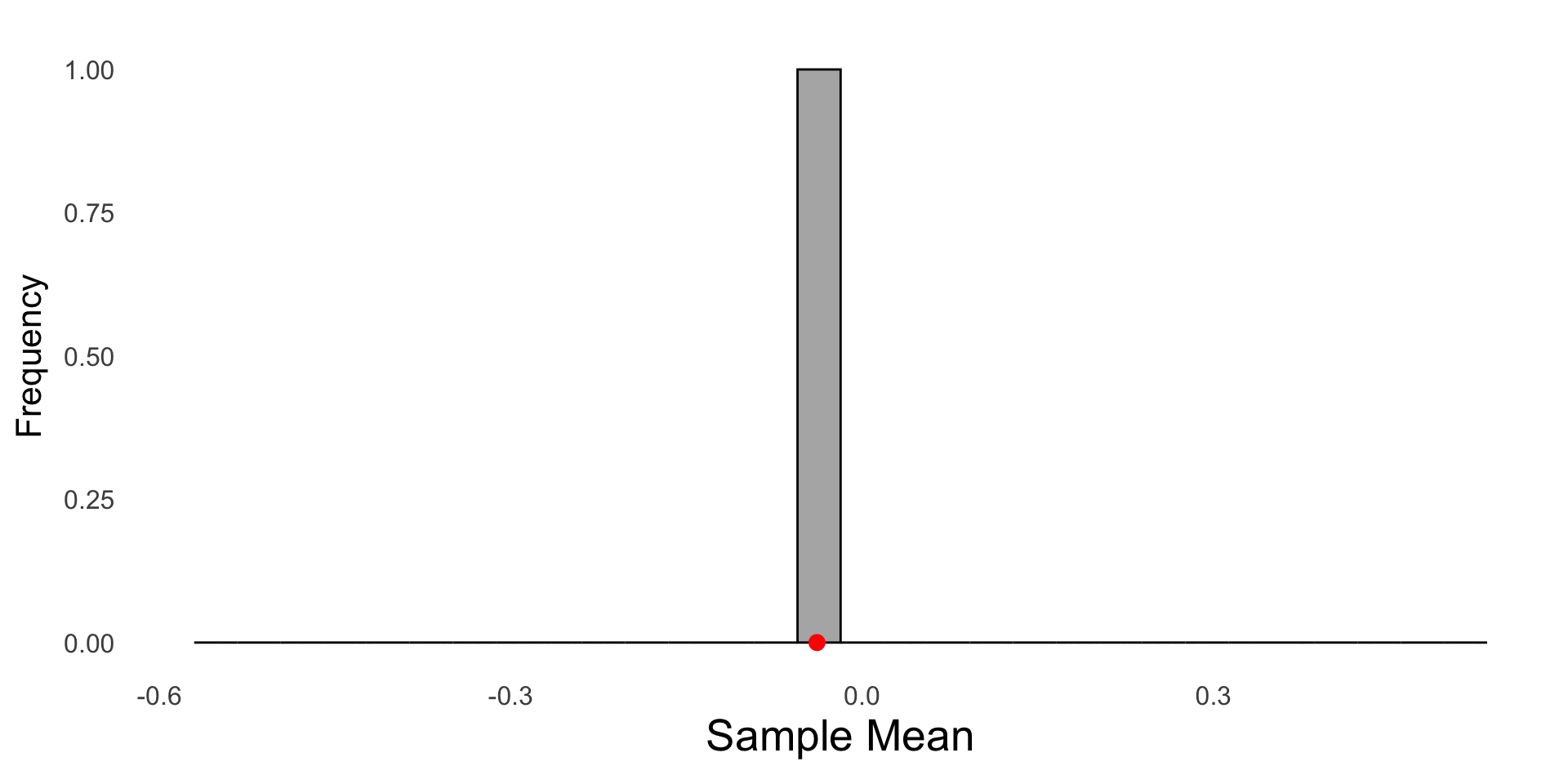
The sample mean from 1000 random samples drawn from a standard normal N(0,1) are saved and plotted in a histogram. Each red dot represents the sample mean of a random sample of size 30 from the population.
Simulated vs Theoretical
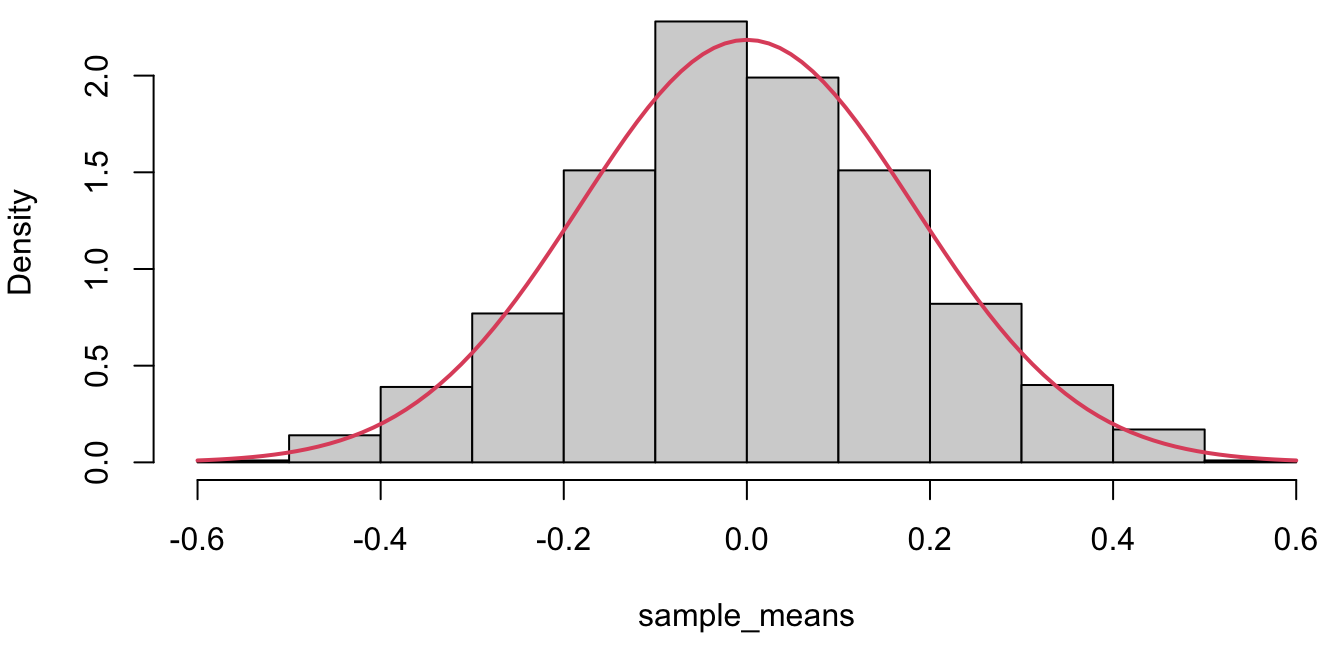
A histogram of 1000 sample means; each \(\bar{X}\) was calculated from a (different) random sample of size 30 drawn from a N(0,1) population. The red curve represents the theoretical sampling distribution of \(\bar{X} \sim N(0, 1/\sqrt{30})\)
Simulated vs Bootstraped

Left: A histogram of the estimates of \(\mu\) obtained by 1000 simulated data sets from the true population (where \(\mu =0\)). Right: A histogram of the estimates of \(\mu\) obtained from 1000 bootstrap samples from a single data set of size 30.
Bootstrap Confidence Interval
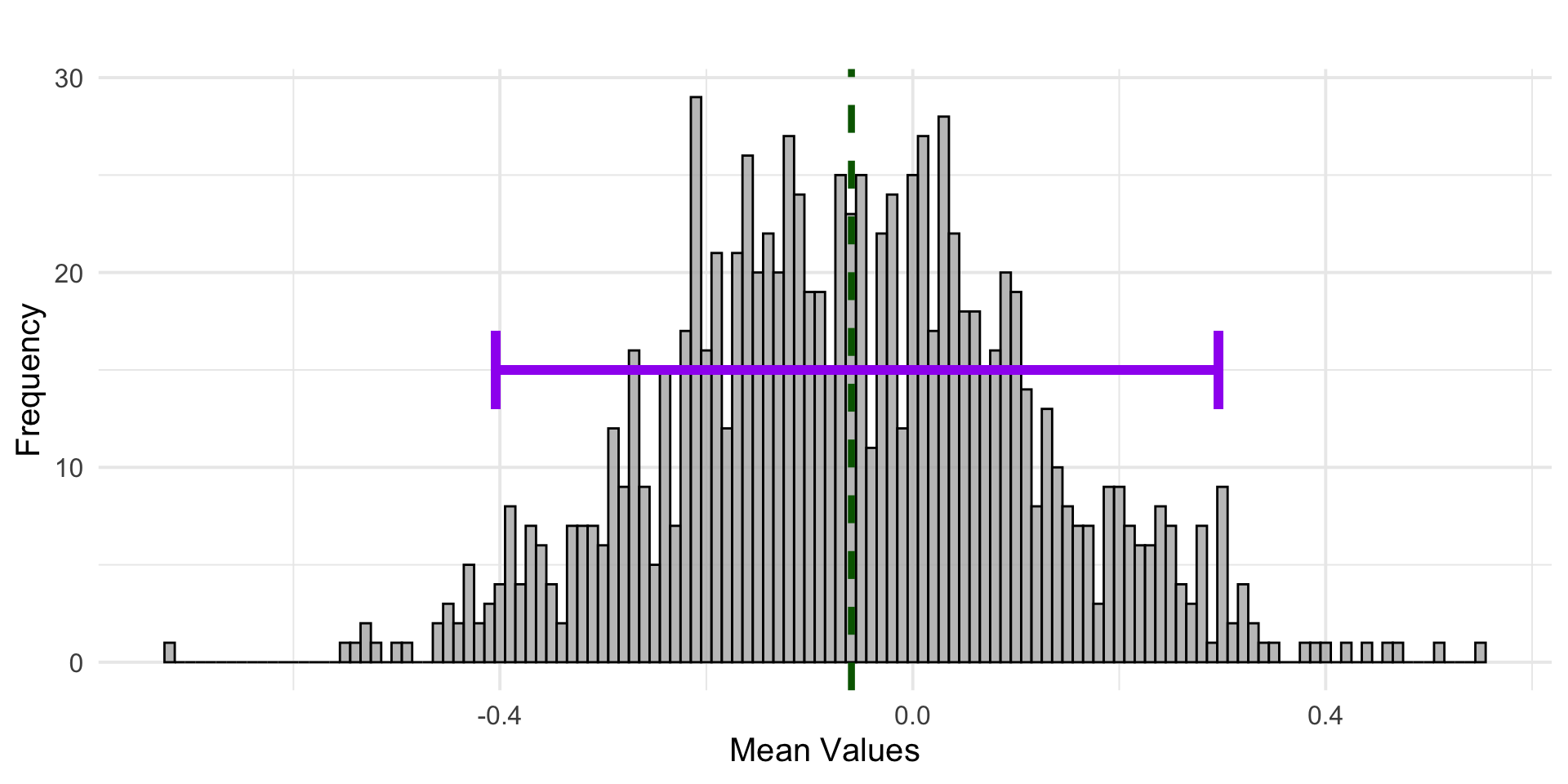
A histogram of the estimates of \(\bar{X}\) obtained by 1000 simulated data sets from the true population (where \(\mu =0\)). The purple indicates the 95% confidence interval as estimated from the bootstrap samples.
Why Bootstrap?
-
The bootstrap used to estimate confidence intervals for parameters, especially when:
- The distribution of the statistic is unknown or complicated.
- The sample size is small, and parametric methods may not be reliable.
More generally, it offers a flexible, non-parametric way to estimate uncertainty for statistical estimates, e.g. standard errors and bias of parameter estimates.
What is Bootstrap?
Definition: Bootstrapping involves repeatedly sampling with replacement from a dataset to create multiple bootstrap samples.
Purpose: It estimates the distribution of a statistic (e.g., mean) by simulating many possible outcomes.
Key Concepts:
- Sampling with replacement vs. without replacement.
- How bootstrapping can be used in machine learning to estimate performance metrics.
Steps
Consider some estimator \(\hat \alpha\). For \(j = 1, 2, \dots B\) 1
- take a random sample of size \(n\) from your original data set with replacement2. This is the \(j\)th bootstrap sample, \(Z^{*j}\)
- calculate the estimated value of your parameter from your bootstrap sample, \(Z^{*j}\), call this value \(\hat \alpha^{*j}\).
Estimate the standard error and/or bias of the estimator, \(\hat \alpha\), using the equations given on the upcoming slides.
Sample with replacement

An illustration of sampling with replacement. The top row shows the original dataset, while the rows below represent different bootstrap samples. Each dot corresponds to a data point, with some points being selected multiple times (overlapping dots) and others not at all.
Bootstrap/full Estimates

Bootstrap Standard Error
Bootstrap estimates the standard error of estimator \(\hat \alpha\) using1
\[\begin{equation} \hat{\text{SE}}(\hat \alpha) = \sqrt{\frac{\sum_{i=1}^{B} (\hat \alpha^{*i} - \overline{\hat \alpha})^2}{B -1}} \end{equation}\]
where \(\overline{\hat \alpha} = \sum_{j = 1}^B \hat \alpha^{*j}\) is the average estimated value over all \(B\) bootstrap samples.
Bootstrap Bias
The bootstrap estimates the Bias of estimator \(\hat \alpha\) using:
\[\begin{align} \hat{\text{Bias}}(\hat \alpha) = \overline {\hat \alpha} - \hat \alpha_f \end{align}\]
where \(\overline{\hat \alpha} = \sum_{j = 1}^B \hat \alpha^{*j}\) and \(\hat \alpha_f\) is the estimate obtained on the original data set.
Regression Example
We simulate 30 observations from a simple linear regression model with intercept \(\beta_0 = 0\) and slope \(\beta_1 = 2\). That is: \[Y = 2X + \epsilon\] where \(\epsilon \sim N(0, 0.25^2)\).
N.B. For ease of notation in the bootstrap section, I will denote \(\beta_1\) (and \(\hat \beta_1\)) by simply \(\beta\) (and \(\hat \beta\)).
Data Generation
We generate a single sample from our population here:
Summary of Fit
And fit a simple linear regression model:
...
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.05798 0.07773 0.746 0.462
x 1.86641 0.14348 13.008 2.17e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.2213 on 28 degrees of freedom
Multiple R-squared: 0.858, Adjusted R-squared: 0.8529
F-statistic: 169.2 on 1 and 28 DF, p-value: 2.172e-13
...Sampling Distribution of \(\hat \beta_1\)
It can be shown that the sampling distribution of \(\hat \beta_1\) is given by a normal distribution centered at the true value, and standard error given below
\[\begin{align} \hat\beta_1 &\sim \text{Normal}\left(\beta_1, \dfrac{\sigma^2}{S_{xx}}\right) & \text{where } S_{xx} = \sum_{i=1}^n (x_i - \overline x)^2 \end{align}\]
Note that this is an unbiased with known standard error!
Theoretical Bias and SE
In other words our we are using an unbiased estimator for slope (bias = 0) with the following theoretical standard error:
\[\begin{align} SE(\hat\beta_1)& = \sqrt{\dfrac{\sigma^2}{S_{xx}}} = \sqrt{\dfrac{0.25^2}{2.38}} = 0.1621 \end{align}\]
Estimates for a single fit
Notice that our summary output provides an estimate of the standard error of \(\hat \beta_1\) from the summary of the lm output (calculated using):
\[\hat{SE}(\hat\beta_1)=\sqrt{\frac{\sum_i\hat\epsilon_i^2}{(n-2)\sum_i(x_i-\bar x)^2}} = 0.1435\]
Since we’ll need this later, note the following estimate on the original data: \(\hat \beta_1 = 1.8664\) (we’ll call this \(\hat\beta_f\) later)
Bootstrap Simulation
While it is a fairly routine to arrive at these equations, this might not be the case for more complicated scenarios.
As an exercise, let’s pretend that we do not know what the standard error and bias of our estimator \(\hat \beta_1\) is.
Instead, let’s estimate the bias (which we know should be 0 since \(\hat \beta_1\) is and unbiased estimator) and standard error using the bootstrap method.1
R code
set.seed(311)
bootsamp <- list()
bootsmod <- list()
bootcoef <- NA
B = 1000 # number of bootstrap samples to be taken
for(i in 1:B){
Zj <- xy[sample(1:n, n, replace=TRUE),]
bootsamp[[i]] <- Zj # store our bootstrap samples in a list
bootsmod[[i]] <- lm(Zj[,2]~Zj[,1])
bootcoef[i] <- bootsmod[[i]]$coefficient[2] # ^betaj
}Bootstrap Estimate for Bias
We now have \(B = 1000\) estimates for \(\beta\) stored in bootcoef = \(\{\hat \beta^{*1},\hat \beta^{*2}, \dots, \hat \beta^{*1000}\}\) and the following values
\[\begin{gather} \overline{\hat \beta} = \sum_{j = 1}^{B = 1000} \hat \beta^{*j} = 1.8704 \quad \hat \beta_f = 1.86641 \\ \text{Bias}(\hat \beta) = \overline {\hat \beta} - \hat \beta_f = 1.8704 - 1.8664 = 0.004 \end{gather}\]
where \(\beta_f\) is obtained from the summary of fit on the full (i.e. original unbootstraped) data.
Bootstrap Estimate for SE
And the estimate for SE is given by this:
[1] 0.1543347[1] 0.1543347\[\begin{align} \hat{\text{SE}}(\hat \beta) &= \sqrt{\frac{\sum_{i=1}^{B} (\hat \beta^{*i} - \overline{\hat \beta})^2}{B -1}} = 0.1543 \end{align}\]
Bootstrap vs Theoretical SE(\(\hat\beta_1\))
The bootstrap estimate (=0.1543) is closer to the theoretical value (=0.1621) that the estimate obtained in the lm summary table from the (single) original data set (= 0.1435).
Important
This estimate was created with no knowledge of the theoretical sampling distribution!!
Had we not known the theoretical sampling distribution for \(\hat \beta_1\) the bootstrap enables us to construct confidence intervals, perform hypothesis test (calculate \(p\)-values!)
Bootstrap vs Simulation-based SE(\(\hat\beta_1\))
- For fun, let’s compare this estimate to the simulation-based method which allows us to resample from this population many many times, …
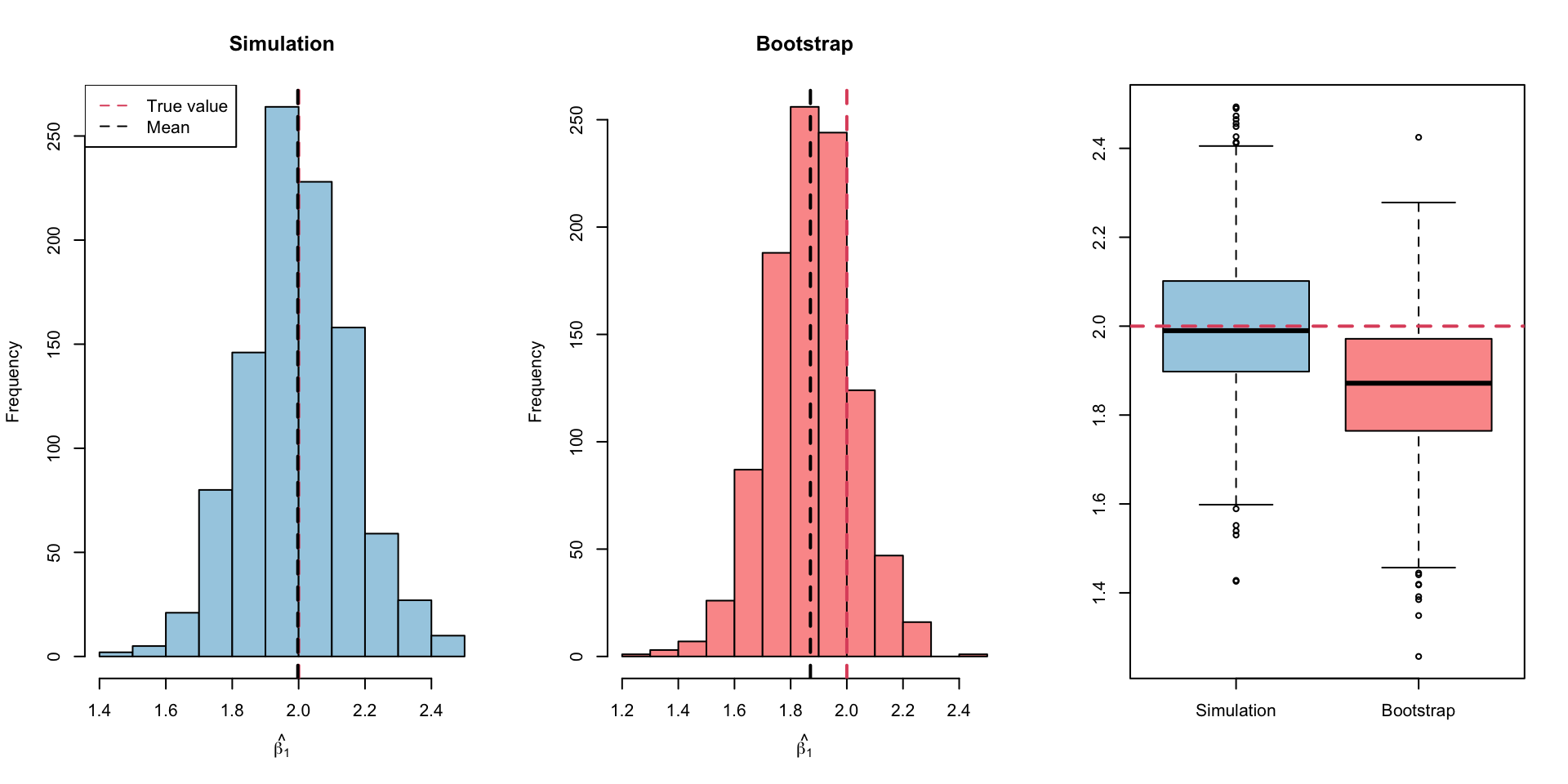
Results (Visual)
Results (Table)
SE estimates from the simulation and the bootstrap simulation are very similar to each other and the theoretical value (=0.162)
| Estimate/ R Code | Description |
|---|---|
0.143 = from output table summary(fullfit)
|
Estimate from a single fit on the full data |
0.154 = sd(bootcoef)
|
Bootstrap estimate obtained from 1000 bootstrap samples |
0.160 = sd(coefs)
|
Standard errors obtained from resampling from the population 1000 times. |
iClicker
Resampling with CV vs Bootstrap
What is the primary difference between the resampling technique used in cross-validation (CV) and the bootstrap method?
- In CV, samples are drawn with replacement, while in the bootstrap method, samples are drawn without replacement.
- In CV, samples are drawn without replacement, while in the bootstrap method, samples are drawn with replacement.
- CV divides the dataset into distinct subsets (folds), while the bootstrap resamples with replacement from the original dataset.
- CV divides the dataset into distinct subsets (folds), while the bootstrap resamples without replacement from the original dataset.
iClicker
Goals of CV vs Bootstrap
What is the primary difference in the goals of CV and the bootstrap method?
- CV aims to estimate the variability of a statistic, while the bootstrap is used to estimate the model’s predictive performance.
- CV focuses on estimating model performance by minimizing overfitting, whereas the bootstrap is used to estimate the variability of a statistic or parameter.
- Both CV and the bootstrap are used for estimating the model’s predictive performance.
- CV and the bootstrap both aim to reduce the bias of an estimator.
Computing Bootstrap CI
We alluded to the potential of computing confidence intervals (CI) with the bootstrap.
We already saw how the \(B\) bootstrap estimates of parameter \(\alpha\) provide an empirical estimate of the sampling distribution of estimator \(\hat \alpha\).
To obtain confidence intervals from this empirical distribution, we would simply finding appropriate percentiles.
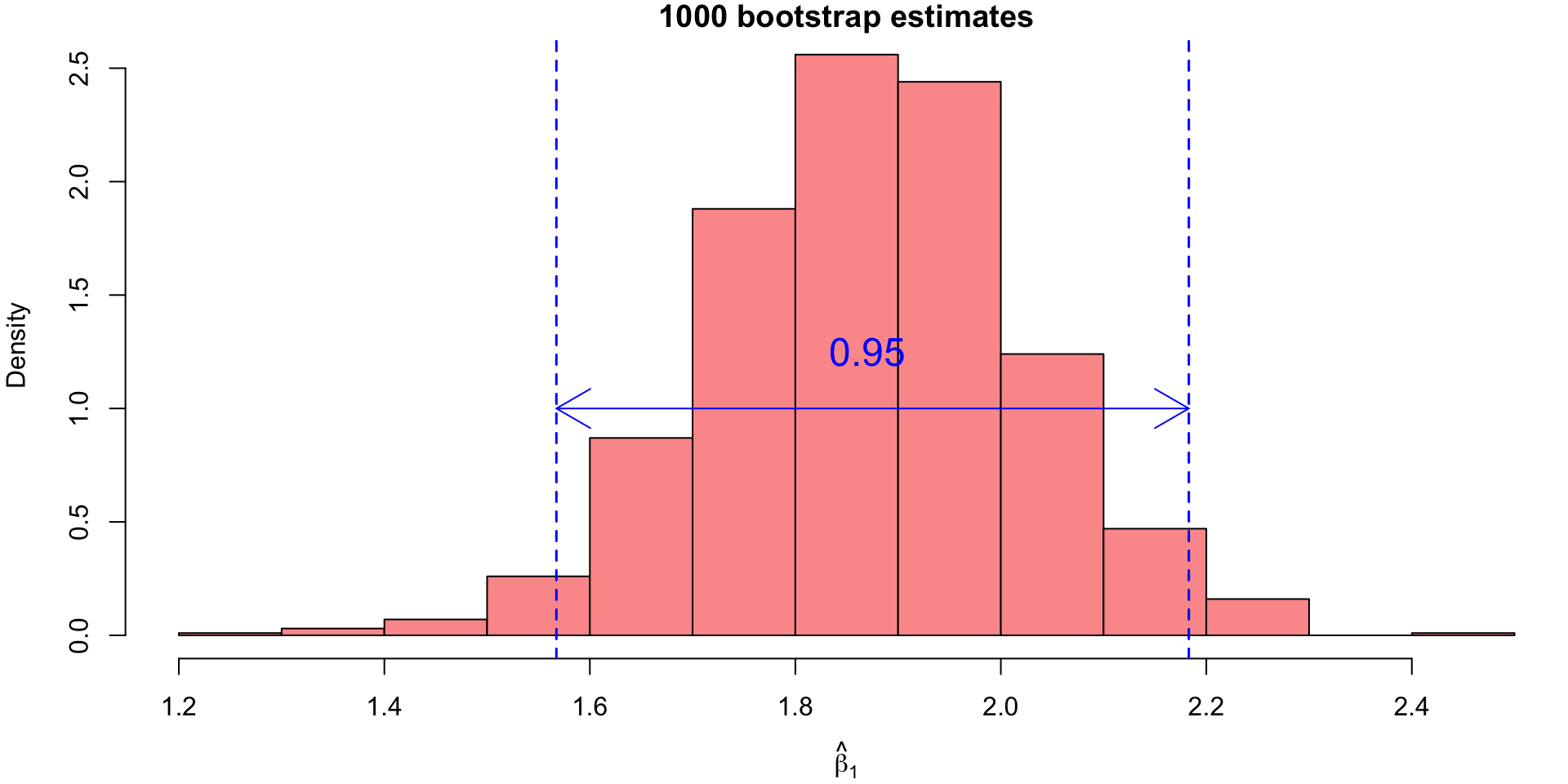
Bootstrap Confidence Intervals (CI)
For our example, the 95% bootstrap CI for \(\beta_1\) would correspond to the 25th value and the 975th value of the sorted bootstrap estimates.
quantile
These values can be found using the quantile() function:
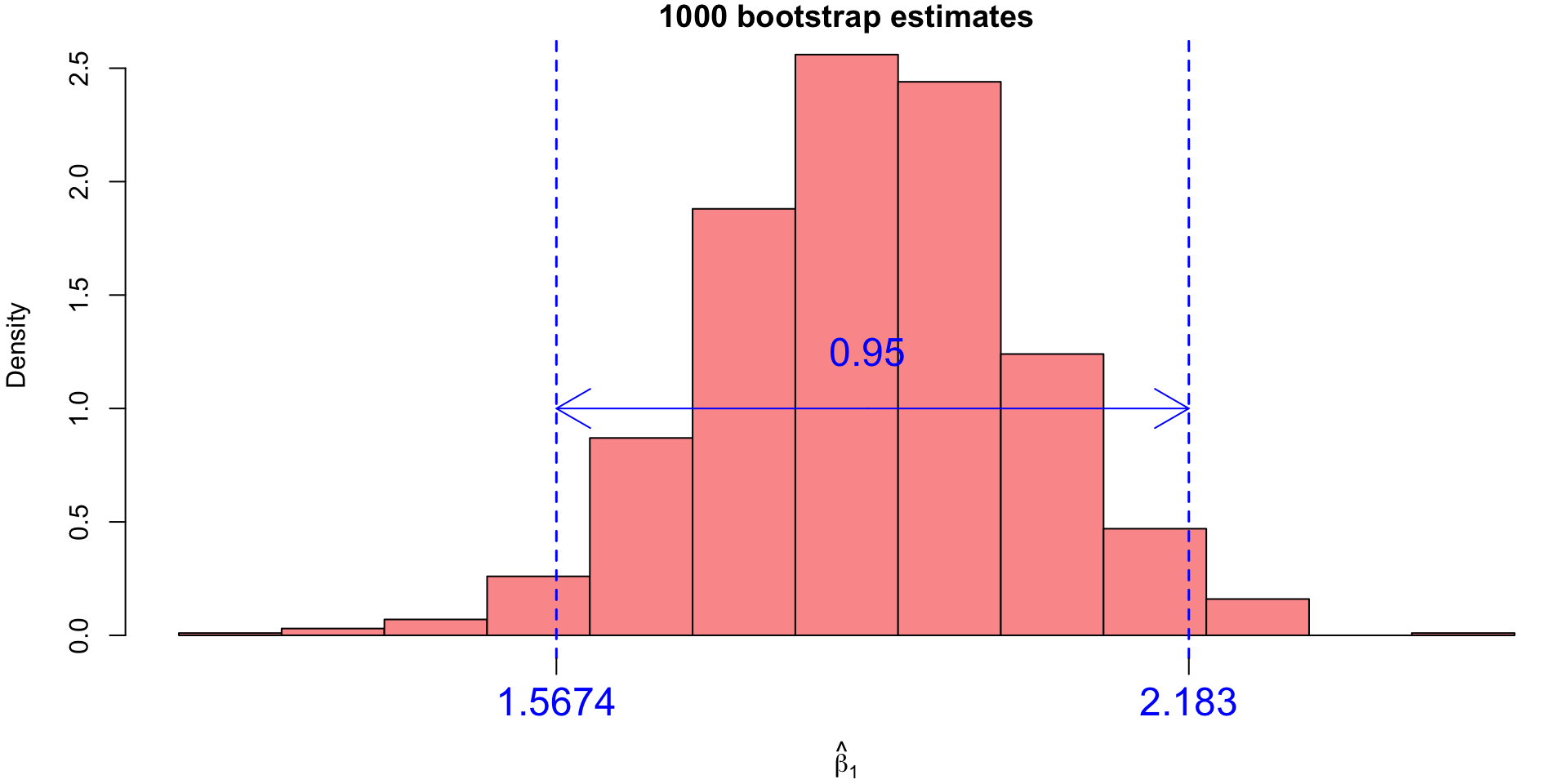
2.5%
1.567437 97.5%
2.182977 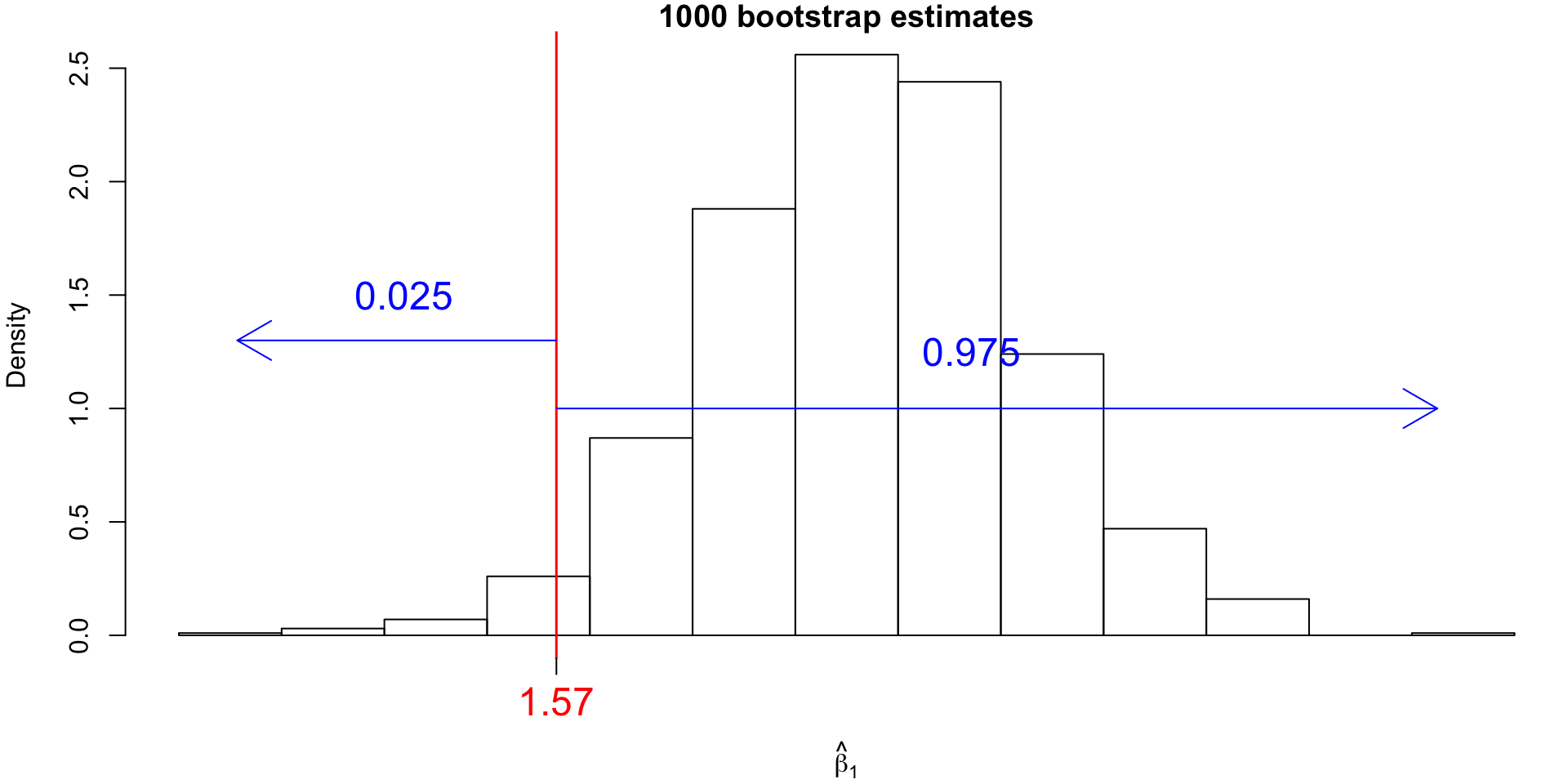
Estimated CI
So we have a 95% CI for true \(\beta_1\) lying within the interval: \[(1.5674, 2.183)\]
Quote
Statistical ‘rock star’ wins coveted international prize
The bootstrap is rarely the star of statistics, but it is the best supporting actor (Dr. Bradley Efron)
You can hear Dr. Brad Efron speak about the bootstrap here.
Dr. Efron gives a nice interview with the ILSR authors here.
Comments
Note that the center of the bootstrap histogram is centered at \(\hat \beta_f\), the estimate obtained using the full data, rather than at \(\beta_1 = 2\) (the true simulated value).
While we will use the bootstrap to estimate the SE of our estimator, typically the point estimate will be given by the estimate obtained using the full data, rather than say the mean of the bootstrap estimates.