Data 311: Machine Learning
Lecture 7: Bayes Classifier, KNN Classification and Discriminant Analaysis
University of British Columbia Okanagan
Introduction
-
Last class we introduced the logistic regression model used for binary classification1. Therein we modeled:
\[ \Pr( \text{class} = 1 \mid \text{data}) \]
-
Today we will continue with classification methods:
- Bayes Classifier
- KNN Classification
- [LDA] Linear Discriminant Analysis
Test Error Rate
- A good classifier is one for which the test error is smallest.
- The test error rate associated with a set of test observations of the form (\(x_0, y_0\)) is given by
\[\begin{equation} \text{Avg}(I(y_0 \neq \hat y_0)) \end{equation}\]
where \(\hat y_0\) is the predicted class label that results from applying the classifier to the test observation with predictor \(x_0\).
Bayes Classifier
“Bayes classifier” is a general concept that refers to any classifier that makes predictions based on Bayes theorem
A Bayes classifier calculates the probability of a particular class given a set of features and then selects the class with the highest probability as the prediction.
It can be shown1 that the Bayes Classifier minimizes the testing error given on the previous page.
Bayes Theorem
Bayes’ theorem relates conditional probabilities.
\[\begin{equation} P(A|B) = \frac{P(B|A) \cdot P(A)}{P(B)} \end{equation}\]
\(P(A|B)\) is the conditional probability of event A occurring given that event B has occurred.
\(P(B|A)\) is the conditional probability of event B occurring given that event A has occurred.
\(P(A)\) is the marginal probability of event A
\(P(B)\) is the marginal probability of event B
Definition
The Bayes classifier assigns each observation to the most likely class, given its predictor/input values (i.e. \(X\)s ).
That is, for some predictor vector \(x_0=(x_{01}, x_{02}, \ldots, x_{0p})\) we should assign the class \(j\) where the following conditional probability is maximized: \[P(Y=j \mid X=x_0)\]
Bayes Decision Boundary Definition
For the two class problem the Bayesian decision boundary is defined as the line where
\[P(Y=1 \mid X=x_0) = P(Y=2 \mid X=x_0)\]
This boundary separates the feature space into two decision regions:
- Region 1 points for which \(P(Y = 1 \mid X = x_0) > 0.5\) and are assigned to class 1
- Region 2 points for which \(P(Y = 1 \mid X = x_0) < 0.5\) and are assigned to class 2
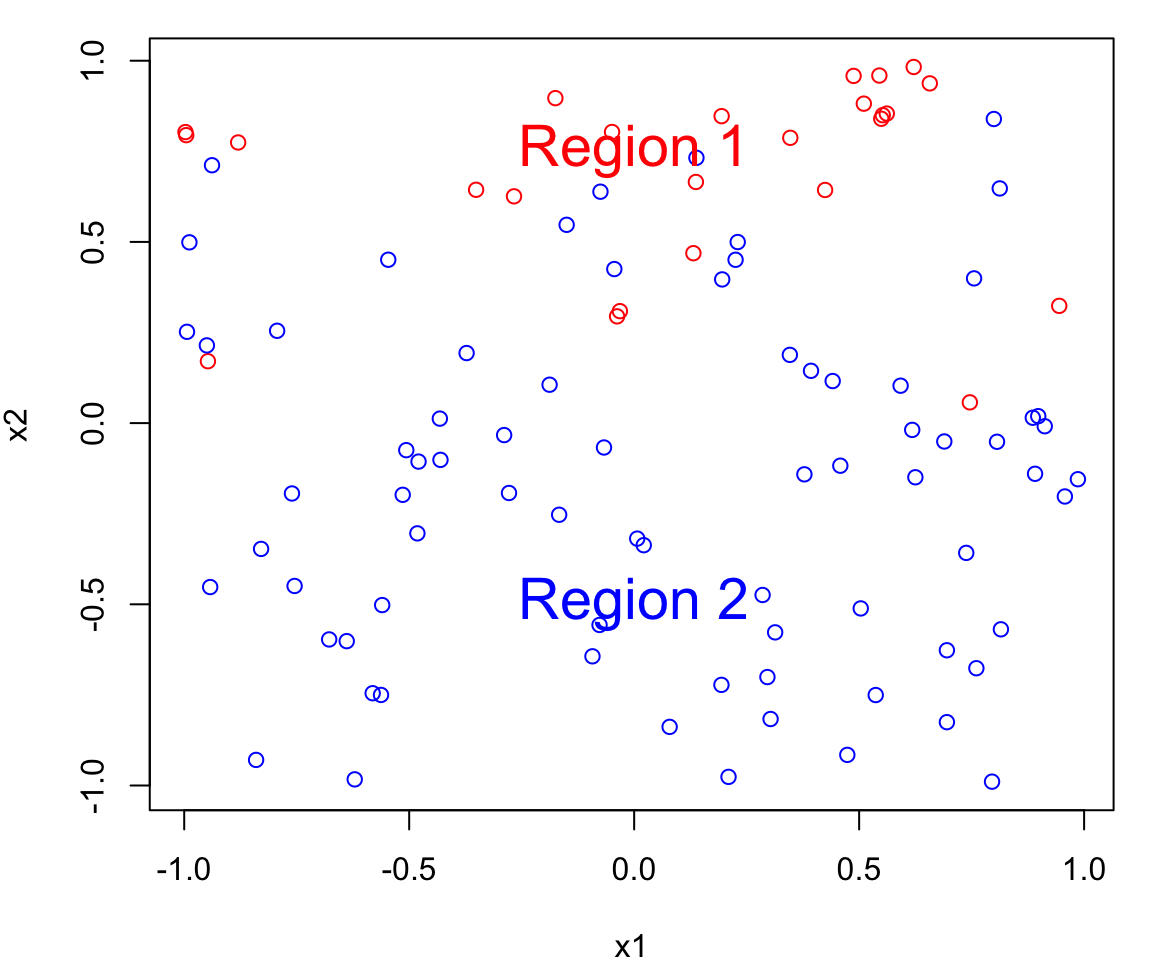
Bayes Classifier Example
Let’s simulate a data set with of two continuous predictors \(X_1\) and \(X_2\); both uniformly distributed from \([-1, 1]\).
-
We create two classes:
- a red group (corresponding to \(Y=0\)) and
- a blue group (corresponding to \(Y=1\))
To generate the class assignment we will only using \(X_2\).
That is \(X_1\) is represents a useless predictor.
Class generation
We will assign an observation to a class based on the following:
If \(x_2 > 0\)
\[\begin{align} P(Y = 0 \mid X_2 = x_2) &= x_2\\ P(Y = 1 \mid X_2 = x_2) &= 1 - x_2 \end{align}\]
If \(x_2 \leq 0\)
\[\begin{align} P(Y = 0 \mid X_2 = x_2) &= 0\\ P(Y = 1 \mid X_2 = x_2) &= 1 \end{align}\]
Notice that \(x_{1i}\) does not provide any information about the class.
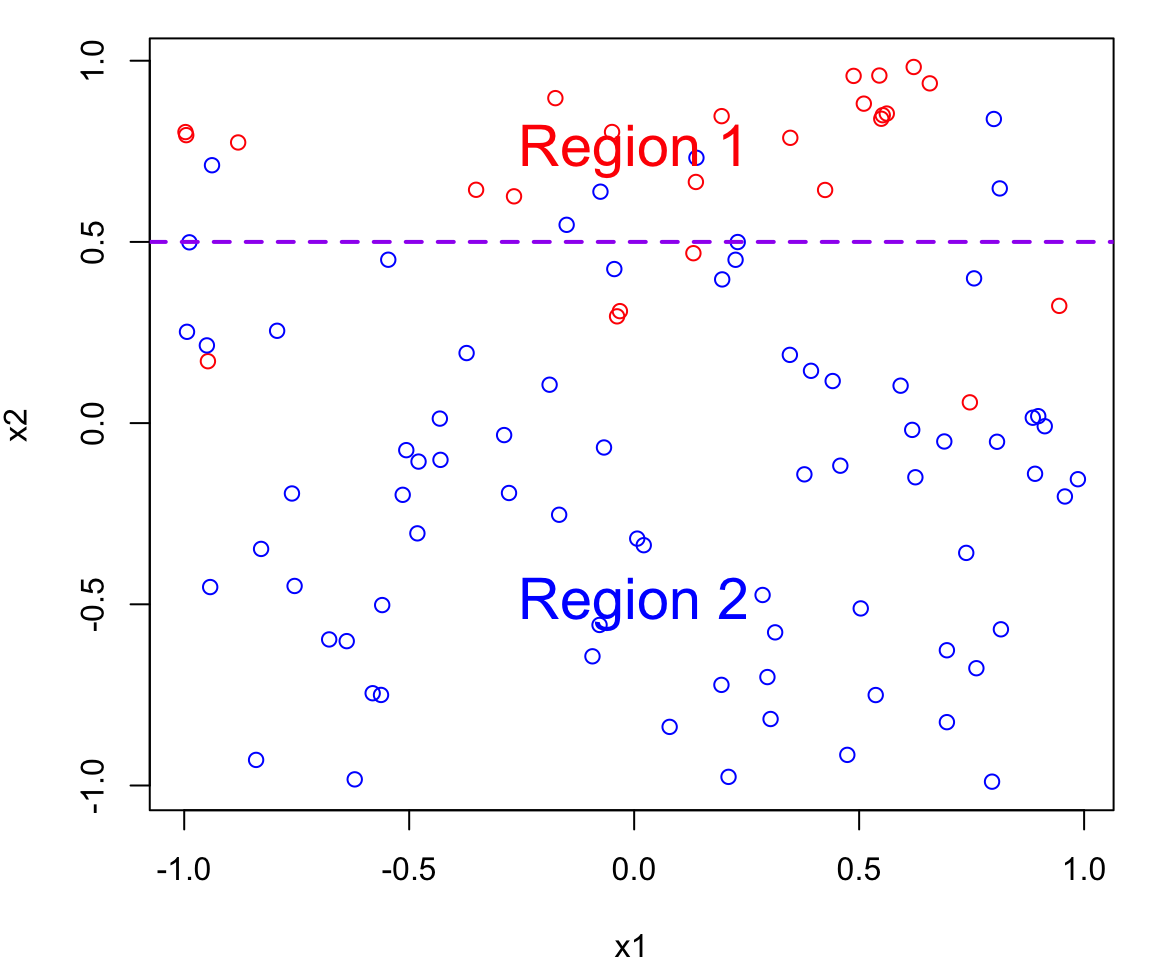
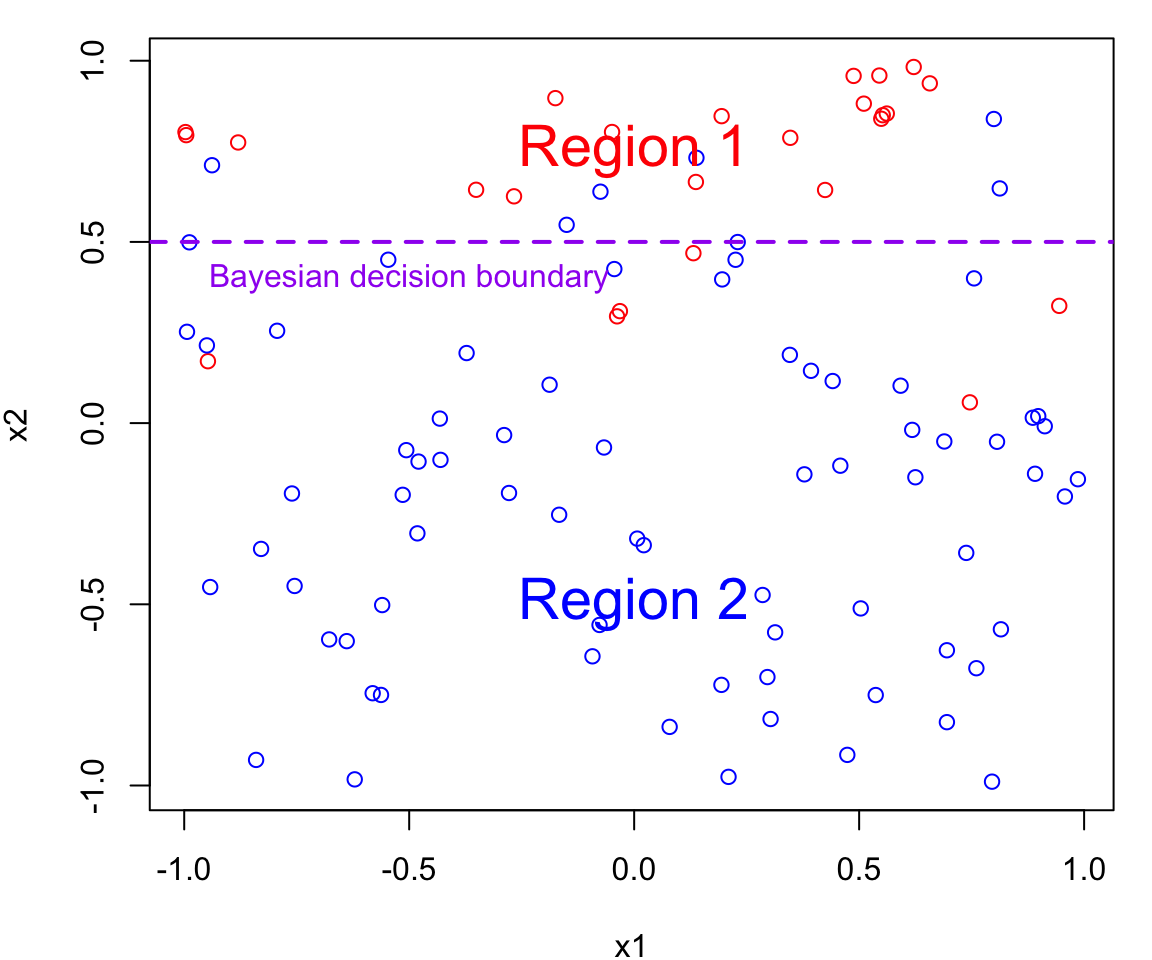
Bayes Decision Boundary
The Bayes decision boundary represents the dividing line in the feature space (i.e. in terms of \(X\)s) that determines how data points are classified into different classes.
In our case data points below the decision boundary (Region 1) are classified as red, while data points above the decision boundary (Region 2) are classified as blue
Note that errors are still going to be made using this classifier since the process is random (points near the boundary are sometimes going to red in Region 2 and sometimes blue in Region 1)
Optimal Boundary
The Bayes decision boundary represents the optimal decision boundary because it’s based on the true underlying probability distributions of the data.
In other words, the Bayes decision boundary is going to make fewer mistakes than any other classifier you will come up with.
Note
In practice, these distributions are often unknown and need to be estimated from data, which can introduce uncertainty.
Bayes Error Rate
On average, Bayes classifier will yield the lowest possible test error rate given by the following expectation (averages the probability over all possible values of \(X\)) \[1-E\left( \max_j P(Y=j \mid X) \right)\]
The above is called the Bayes error rate which represents the minimum possible error rate that any classifier can achieve for a given classification problem.
From simulations to real data
- In theory we would always like to predict qualitative responses using the Bayes classifier.
- But for real data, we do not know the conditional distribution of \(Y\) given \(X\), and so computing the Bayes classifier is impossible.
- Therefore, the Bayes classifier serves as an unattainable gold standard against which to compare other methods.
Classification Algorithms
Many approaches attempt to estimate the conditional distribution of \(Y\) given \(X\), and then classify a given observation to the class with highest estimated probability.
-
For example, Logisitic Regression modeled
\[ \Pr(Y_i = 1 \mid X_i = x_i) \tag{1}\]
and \(X_i\) is assigned class 1 if this probability is \(\geq 0.5\).
Another example is the \(K\)-nearest neighbors (KNN) \(\dots\)
KNN Classification
Given a positive integer \(K\) and a test observation \(x_0\), the KNN classifier first identifies the \(K\) points in the training data that are closest to \(x_0\), represented by \(\mathcal{N}_0\).
For each class \(j\), find \[\begin{equation} P(Y=j \mid X=x_0) \approx \frac{1}{K} \sum_{i \in N_0} I(y_i = j) \end{equation}\]
Assign observation \(x_0\) to the class (\(j\)) for which the above probability is largest; “ties” are typical broken at random (i.e. a coin toss)
in R
To demo knn classification we will be using the knn function from the class package performs (other alternatives exist)
-
trainmatrix or data frame of training set cases. -
testmatrix or data frame of test set cases. -
clfactor of true classifications of training set -
knumber of neighbours considered.
k Nearest Neighbours Example
Let’s apply this algorithm to the simulated data considered previously.
More specifically, we will fit
knnfor \(k = 20, 15, 9, 3\) and \(1\)Fair warning that the decision boundary and decision regions will not be as clear cut as our previous example.
We will assess the error rate for each (code presented in lab)
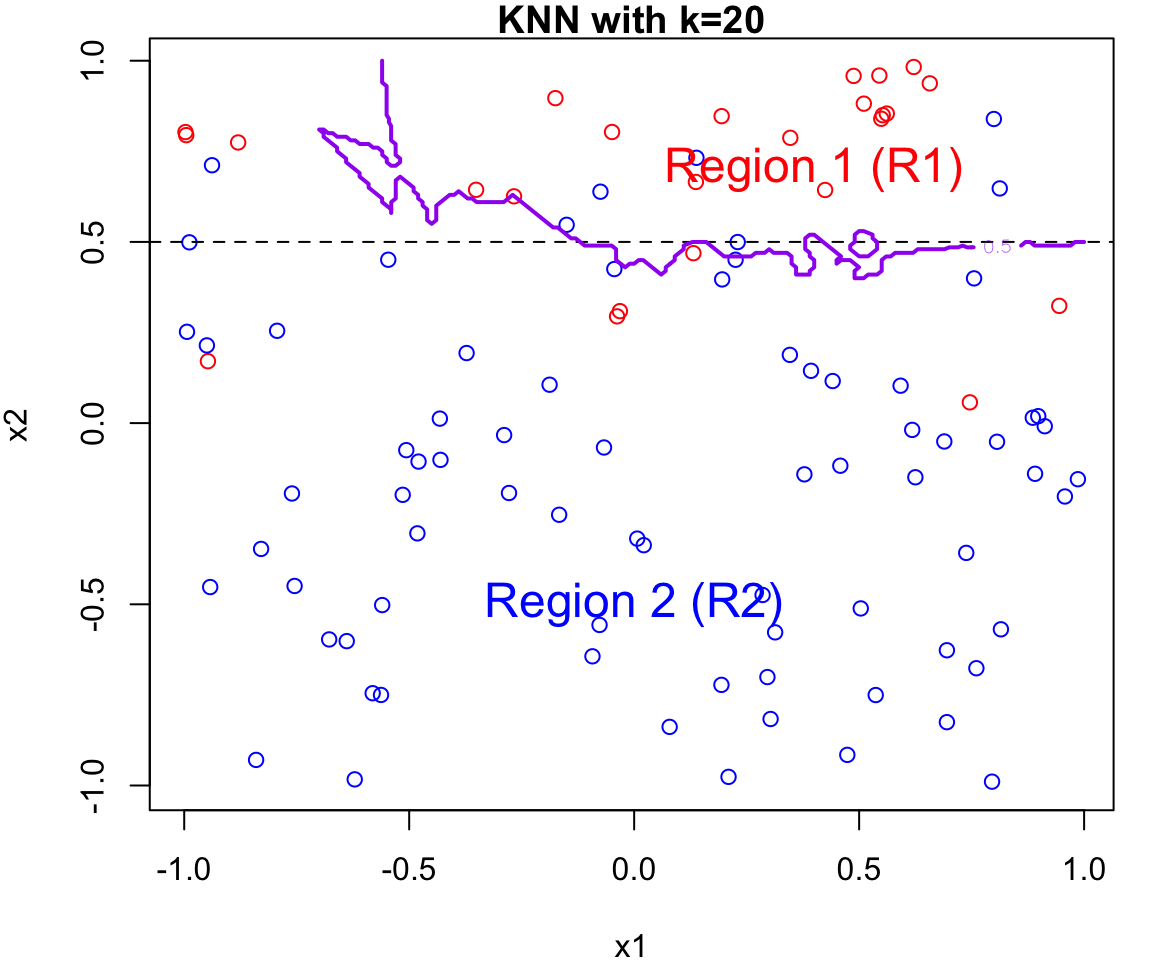
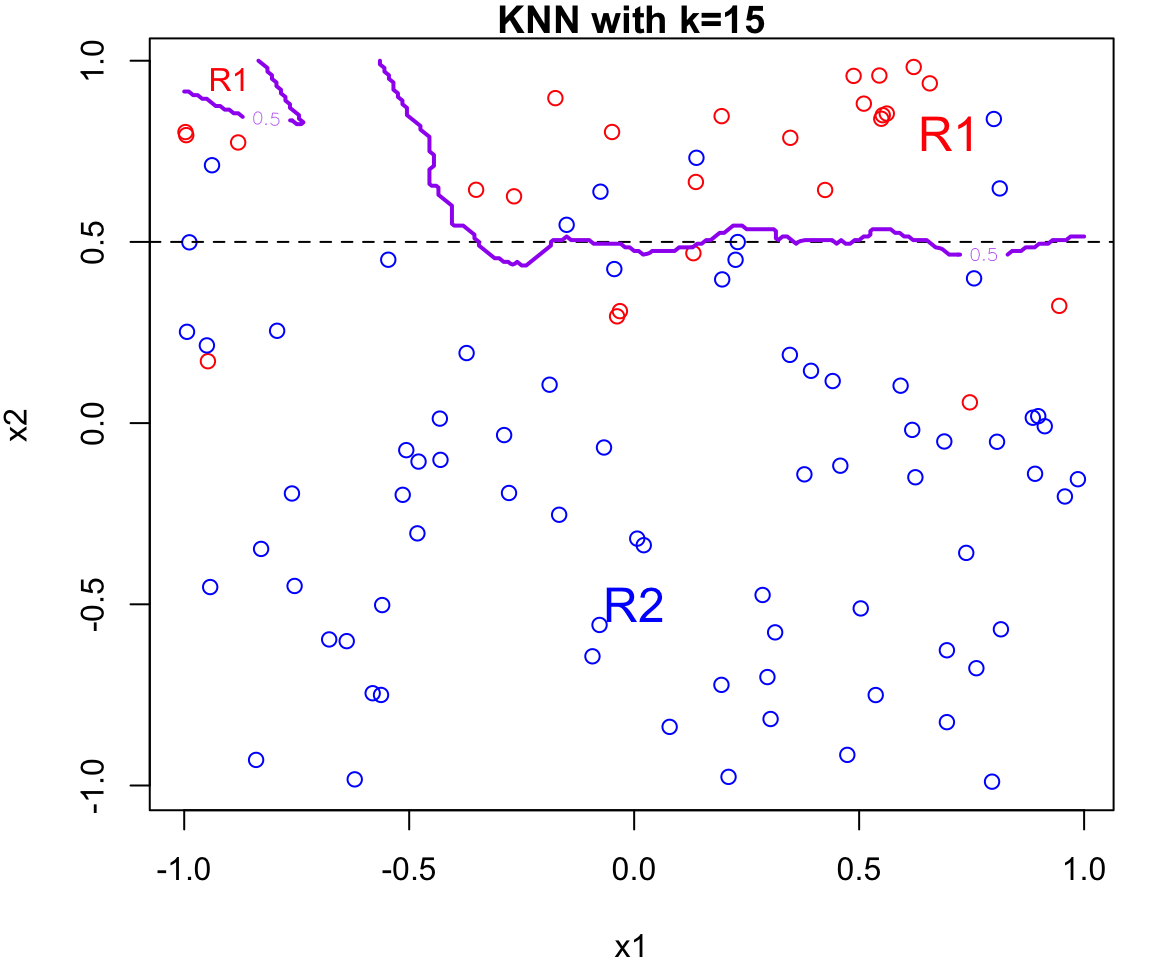
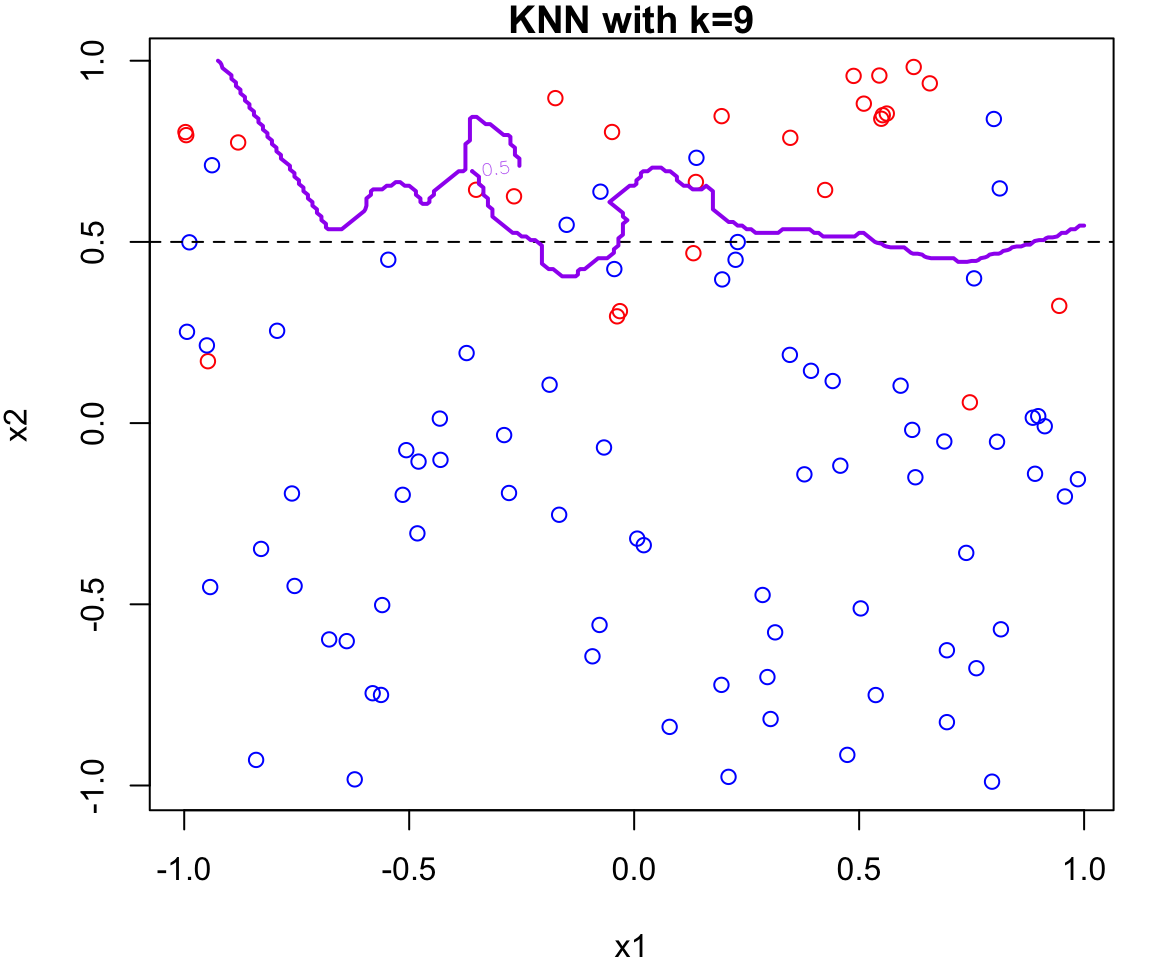
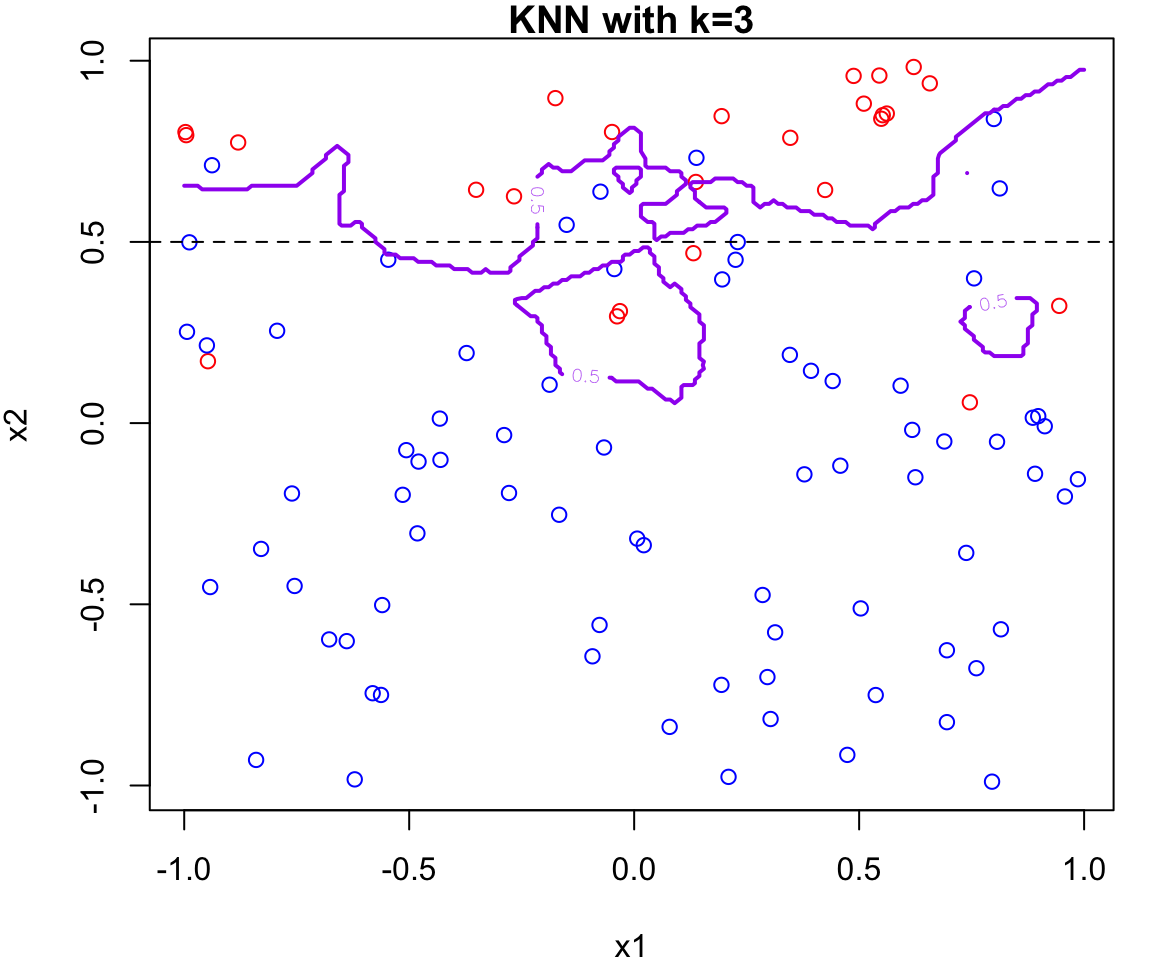
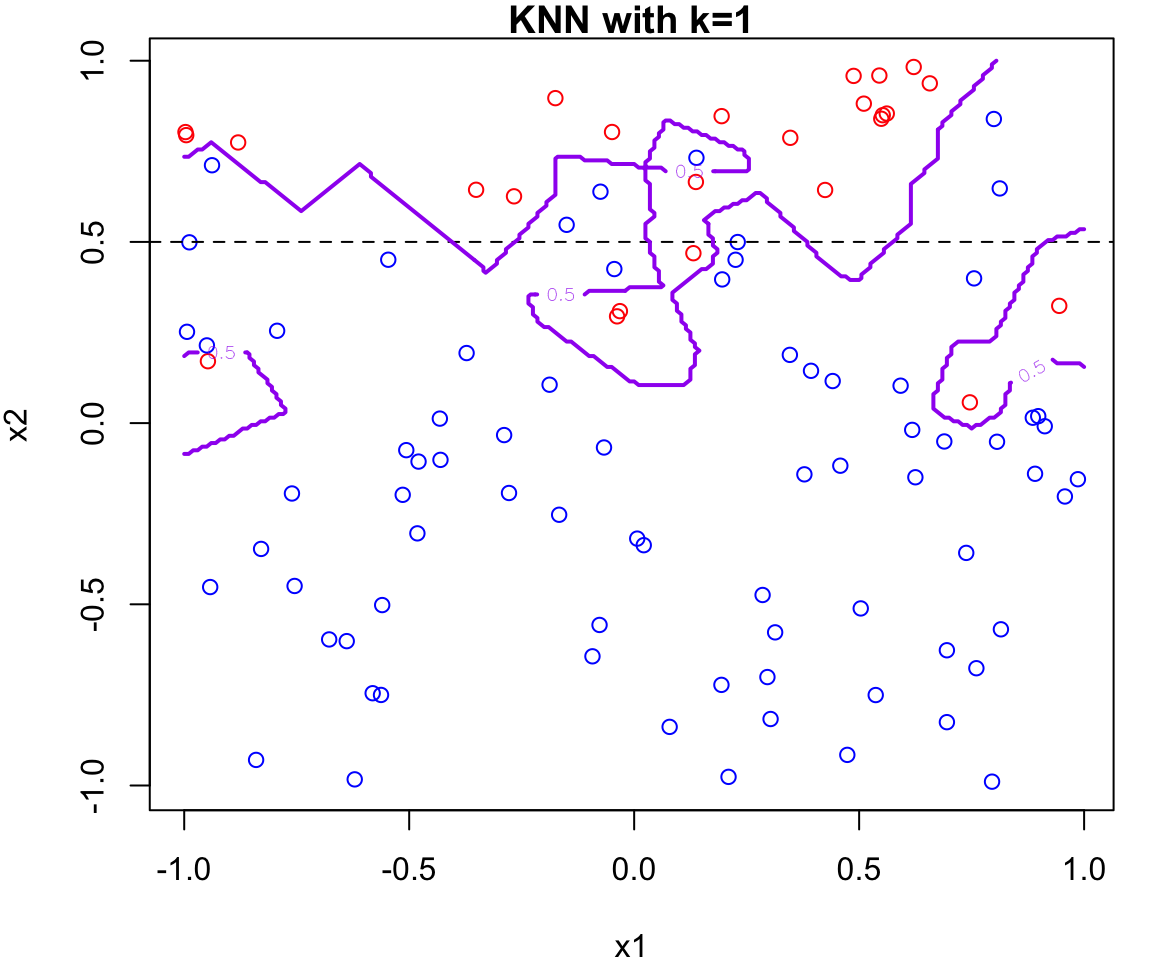
An great interactive demo can be view here
Choosing k
A key component of a “good” KNN classifier is determined by how we choose \(k\), the number of nearest neighbours.
-
\(k\) in a user-defined input which we can set anywhere from 1 to \(n\) (the number of points in our training set)
Choose odd k to avoid ties in binary classification.
Use cross-validation to find the optimal k value for your dataset. (more on this later in the course).
iClicker: small neighborhood
What happens to the decision boundary in KNN when the value of \(k\) is too small (e.g., k=1)?
- The model becomes more robust to noise
- The decision boundary becomes smoother
- The model becomes more sensitive to noise
- The model becomes less flexible
Correct answer: C.
iClicker
k related to bias-variance
In KNN, increasing \(k\) tends to:
- Increase bias and decrease variance
- Increase both bias and variance
- Decrease both bias and variance
- Decrease bias and increase variance
Correct answer: A. Increase bias and decrease variance
Recap
To estimate the conditional distribution of \(Y\) given \(X\),
KNN did this using a “voting” system with its nearest neighbours \(\Pr(Y=j \mid \boldsymbol x) \approx \frac{1}{K} \sum_{i \in N_0} I(y_i = j)\)
Logistic regression uses the logistic function, e.g. \[\Pr(Y = 1 \mid \boldsymbol x) = \frac{e^{\beta_0 + \beta_1x}}{1 + e^{\beta_0 + \beta_1x}}\]
Let’s discuss linear and quadratic discriminant analysis as alternative methods for approximating the conditional distribution of \(Y\) given \(X\) \(\dots\)
Bayes Theorem
\[ \Pr(Y = k \mid \boldsymbol x) = \frac{\Pr(\boldsymbol x \mid Y = k)\cdot \Pr(Y = j)}{\sum_{j} \Pr(\boldsymbol x \mid Y = k)\cdot \Pr(Y = k)} \]
we can assume some model for \(\Pr(\boldsymbol x \mid Y = k) \sim f_k(\boldsymbol x)\) (e.g. Gaussian)
\(\Pr(Y=k)\) is the prior, denoted \(\pi_k\). It represents the prior probability of observation \(\boldsymbol x\) belonging to class \(k\)
Posterior Probabilities
Then Bayes’ theorem states that
\[\begin{equation} \Pr(Y = k \mid X = x) = \frac{\pi_k f_k(x)}{\sum_{l=1}^{K} \pi_l f_l(x)} \end{equation}\]
This represents the posterior probability that an observation \(X = x\) belongs to the \(k\)th class.
To estimate \(p_k(x)\), and thereby approximate the Bayes classifier, we can need to estimate the \(\pi_k\)s and \(f_k(x)\).
Discriminant Analysis
We assume a normal (Gaussian) distributions with mean vector \(\boldsymbol{\mu}_k\) and covariance matrix \(\boldsymbol{\Sigma}_k\) for each class
\[f_k(\mathbf x) = \frac{1}{(2\pi)^{p/2}|\boldsymbol{\Sigma}_k|^{1/2}} e^{-\frac{1}{2}(\mathbf{x}-\boldsymbol{\mu}_k)^T\boldsymbol{\Sigma}_k^{-1}(\mathbf{x}-\boldsymbol{\mu}_k)}\]
Important
Depending on our assumptions on \(f_k(\boldsymbol x)\), this leads to linear discriminant analysis (LDA) or quadratic discriminant analysis (QDA).

Which class does the new point as indicated by the star belong to?
To answer this, it might be useful to talk about a decision boundary.
This will depend on our priors and parameters.
Generative Models for Classification
For simplicity, let’s suppose we have two classes, that is \(Y\) can take on two possible values \(K = 1\) or \(K = 2\)
Let \(f_k(X) \equiv \Pr(X\mid Y = k)\) denote the pdf of \(X\) for an observation that comes from the \(k\)th class.
Let \(\pi_k\) represent the overall or prior probability that a randomly chosen observation comes from the \(k\)th class.
LDA for one predictor
For simplicity let’s assume \(x\) is univariate so that \(f_k(x)\) be the is univariate Normal distribution with mean \(\mu_k\) and \(\sigma_k^2\) for an observation from the \(k\)th class.
\[\begin{equation} f_k(x; \mu_k, \sigma_k) = \frac{1}{\sqrt{2\pi}\sigma_k} e^{-\frac{(x-\mu_k)^2}{2\sigma_k^2}} \end{equation}\]
Before we estimate \(f_k(x)\), we will first make some assumptions about its form.
The \(\pi\) without a subscript is the constant 3.14159….
Univariate Normal Distribution
Sticking with univariate for now, suppose each group is normally distributed with different means and variances
\[\begin{align*} f_1(x) &\sim N(\mu_1, \sigma_1^2)\\ f_2(x) &\sim N(\mu_2, \sigma_2^2) \end{align*}\]
Plot of two univariate normals
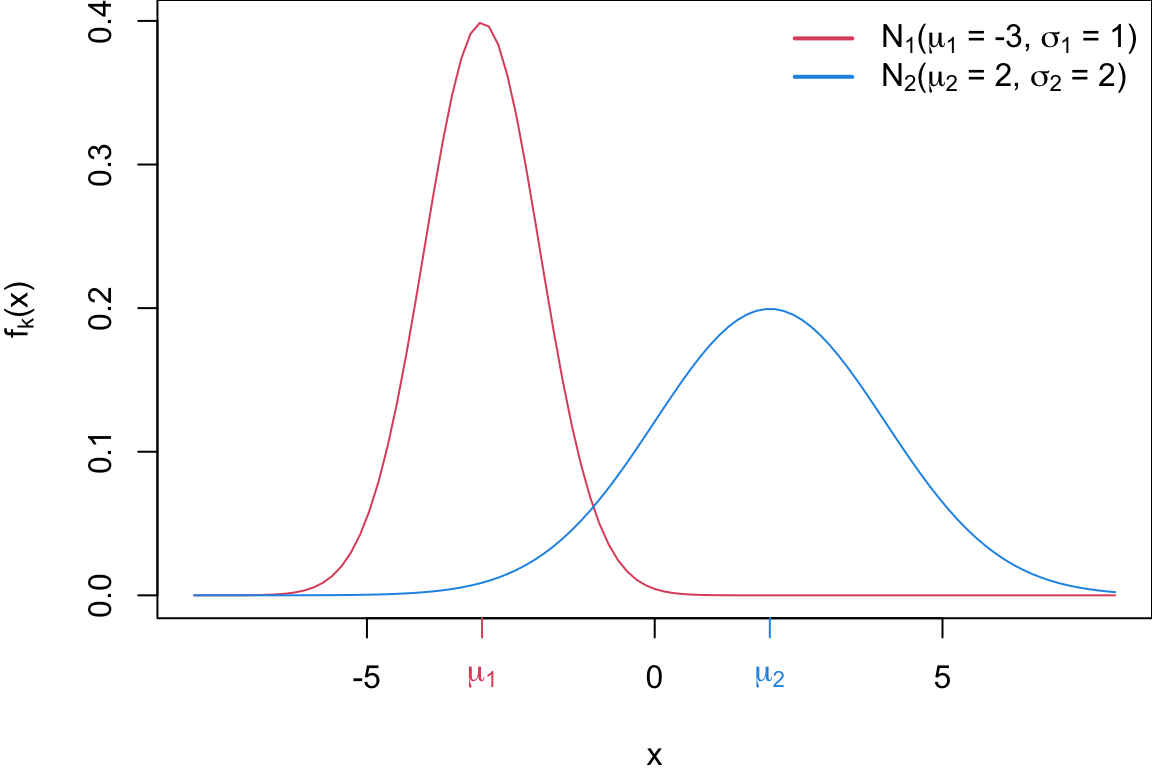
The red curve represents \(f_1 = N(\mu_1 = -3, \sigma_1 = 1)\), and the blue curve represents \(f_2 = N(\mu_2 = 2, \sigma_2 = 2)\).
Code
par(mar = c(3.9,4.1,0,0))
pi1 = 0.6
pi2 = 1 - pi1
curve(dnorm(x, mean = mean1, sd = sd1), col = 2, from = -8, 8, ylab = expression(paste(pi[k]*f[k]*"("* x *")")), lty = 1)
curve(dnorm(x, mean = mean2 , sd = sd2), col = 4, add = TRUE, lty = 1)
gmm <- function(x, pis, mus, sigs){
pis[1]*dnorm(x, mean = mus[1], sd = sigs[1]) +
pis[2]*dnorm(x, mean = mus[2], sd = sigs[2])
}
# curve(gmm(x, c(pi1, pi2), c(mean1, mean2), c(sd1, sd2)), col = 3, add = TRUE)
curve(pi1*dnorm(x, mean = mean1, sd = sd1), col = 2, from = -8, 8, ylab = expression(paste(pi[k]*f[k]*"("* x *")")), lty = 2, add = TRUE)
curve(pi2*dnorm(x, mean = mean2 , sd = sd2), col = 4, add = TRUE, lty = 2)
axis(1, at=mean1, labels = expression(mu[1]), col=2, col.axis=2)
axis(1, at=mean2, labels = expression(mu[2]), col=4, col.axis=4)
ltext <- c(bquote(N[1]*"("*mu[1] *" = "* .(mean1)*", "*sigma[1] *" = "* .(sd1) * ")"),
bquote(N[2]*"("*mu[2] *" = "* .(mean2)*", "*sigma[2] *" = "* .(sd2)* ")"),
bquote(pi[1] *" = "* .(pi1)*", "*pi[2] *" = "* .(pi2)))
legend("topright", lwd= 2, col = c(2,4,NA),
legend = ltext, bty = "n")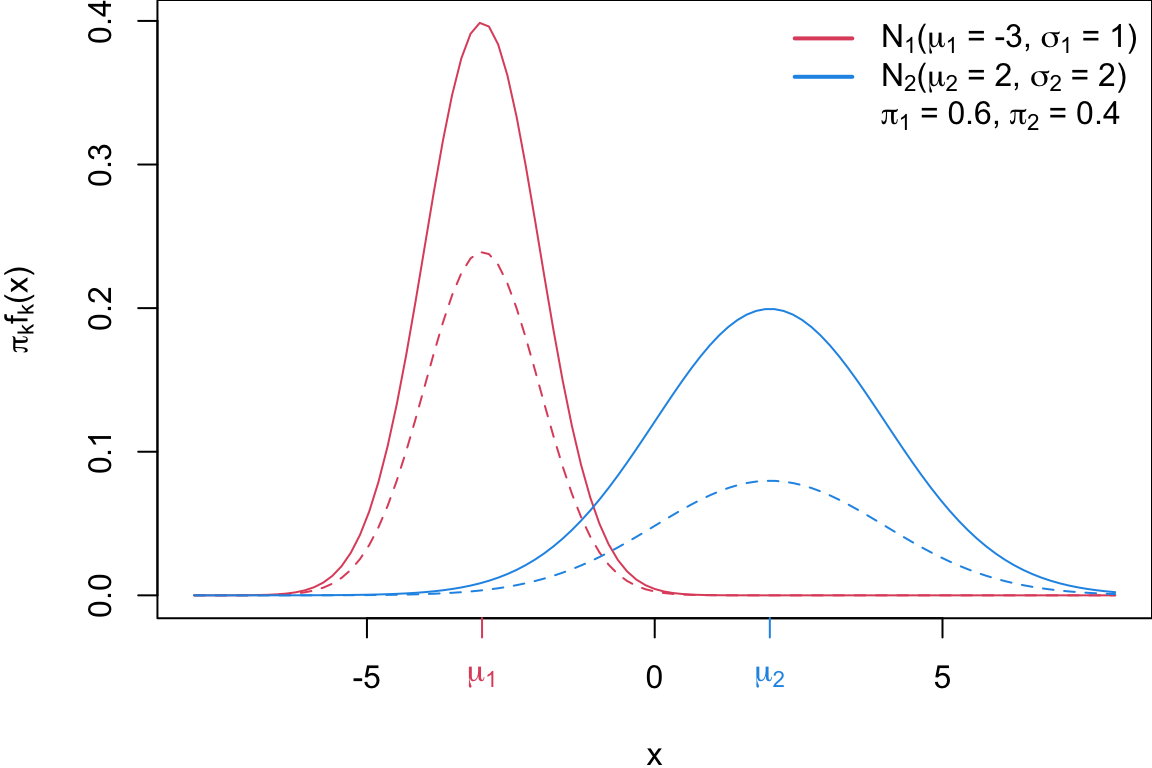
The solid lines denote \(f_k\), the dashed lines show \(\pi_k * f_k\), i.e. the distributions weighted by their class probabilities \(\pi_1 = 0.6\) and \(\pi_2 = 0.4\).
Building a classifier
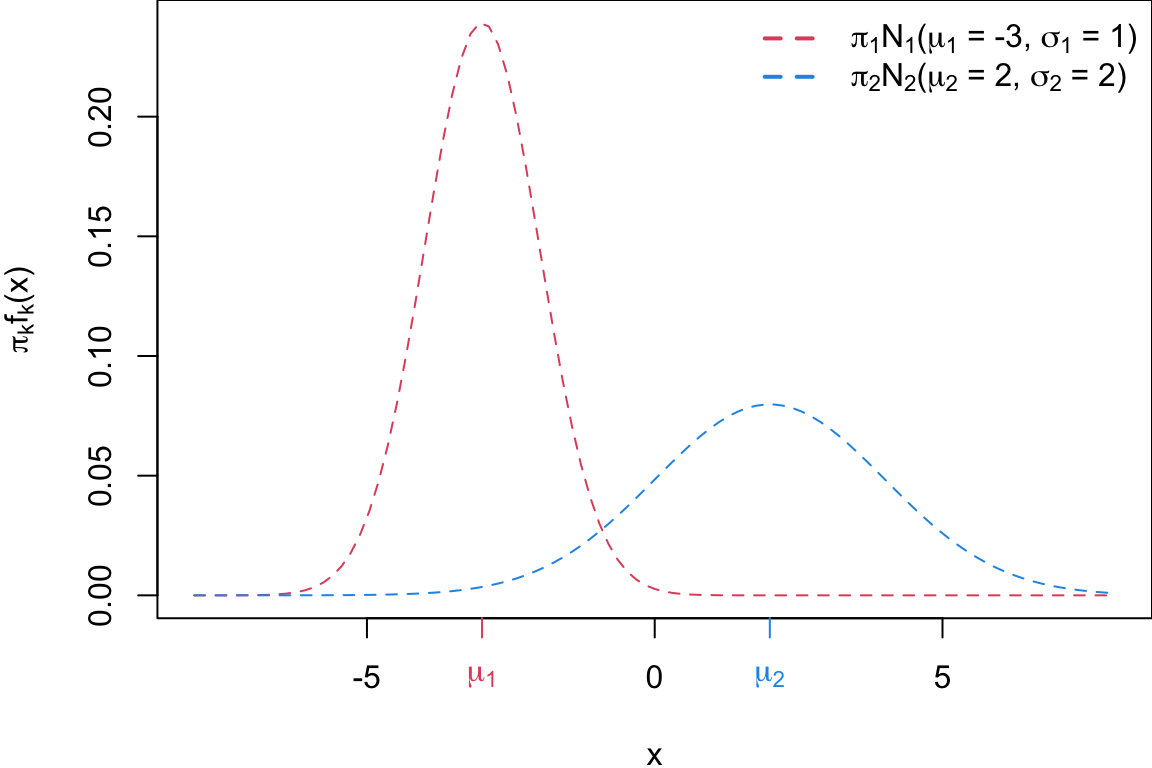
We classify new points according to whichever numerator \((\pi_k f_k(x))\) is the highest.
Question: where would that decision boundary be on this picture?
Case 1
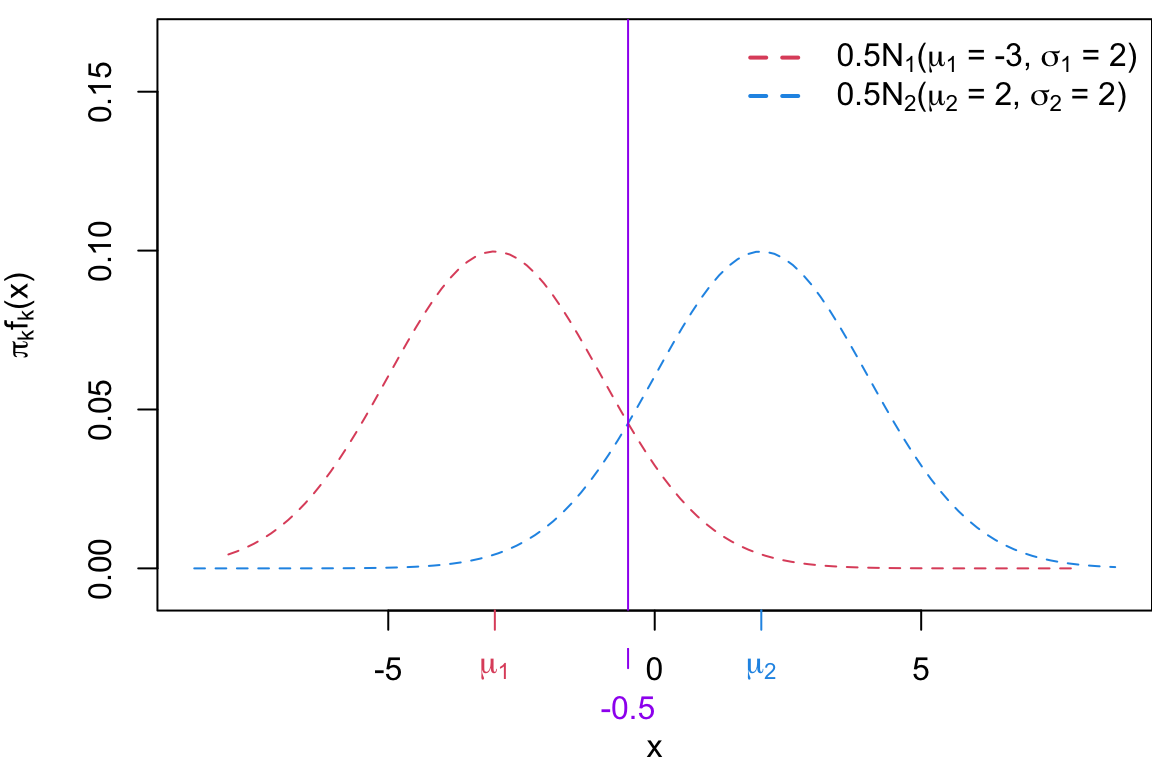
Case 1: \(\sigma_1 = \sigma_2\) and \(\pi_1 = \pi_2\) If the class variances and priors are equal, we have a linear decision boundary which is simply the average of the means: \(x = \frac{\mu_1 + \mu_2}{2}\)
Case 2
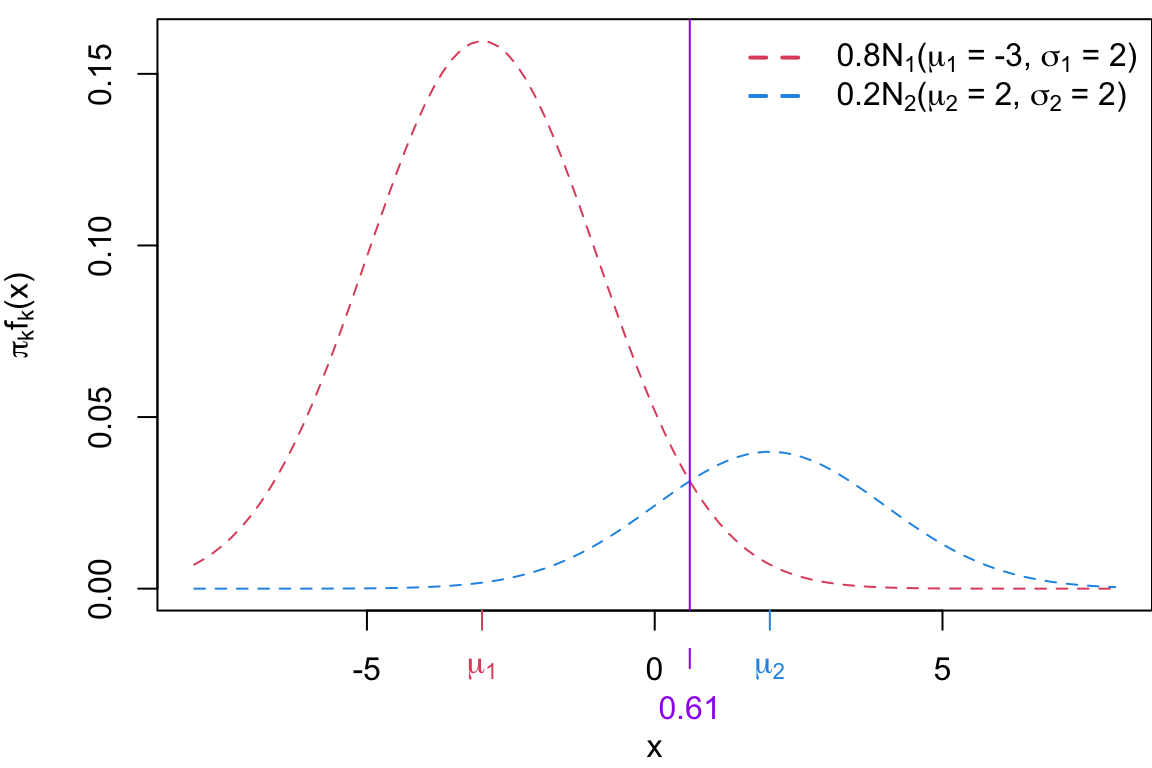
Case 2: \(\sigma_1 = \sigma_2\) and \(\pi_1 \neq \pi_2\) If the class variances are samle and the priors are different, the decision boundary is still linear but shifted toward the class with the lower prior probability (the less likely class), i.e. \(x = \frac{\mu_1 + \mu_2}{2} + \frac{\sigma^2}{\mu_1 - \mu_2} \log \left( \frac{\pi_1}{\pi_2} \right)\)
Case 3
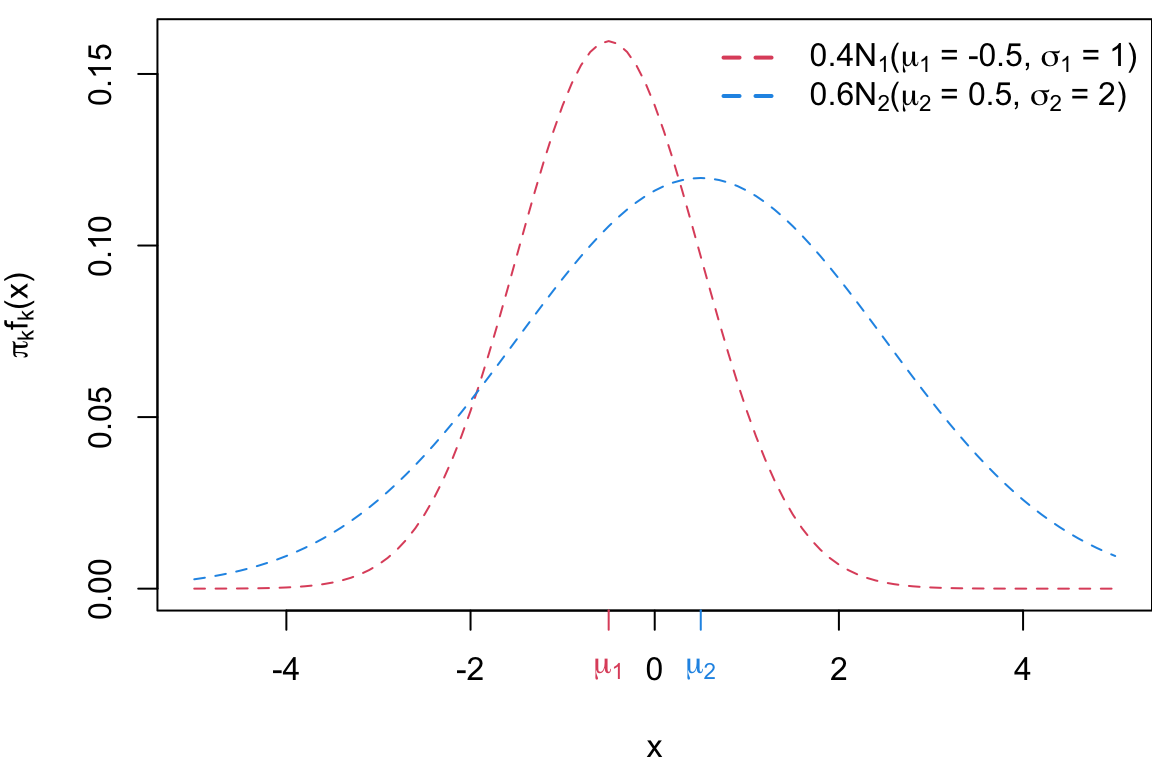
Case 3: \(\sigma_1 \neq \sigma_2\) and \(\pi_1 \neq \pi_2\) If the class variances are samle and the priors are different, the decision boundary is non-linear (quadratic).
LDA vs QDA Assumptions
Linear Discriminant Analysis (LDA) assumes that the variances of all classes are equal, i.e., \(\sigma_1 = \sigma_2 = \sigma\), where \(\sigma\) represents the common variance.
When we relax this assumption and allow for different variances between the groups, we get Quadratic Discriminant Analysis (QDA). In QDA, we have \(\sigma_1 \neq \sigma_2\), meaning the variances differ across the classes.
LDA decision boundary
Since we’re dealing with two classes, we can set the decision boundary where \(p_1(x) = p_2(x)\)
Assumption \(\sigma_1 = \sigma_2\) (but the \(\pi_k\)s will vary)
\[\begin{align} \frac{\pi_1 f_1(x)}{\sum_{l=1}^{K} \pi_l f_l(x)} &= \frac{\pi_2 f_2(x)}{\sum_{l=1}^{K} \pi_l f_l(x)} \\ \frac{\pi_1}{\sqrt{2\pi}\sigma} e^{-\frac{(x-\mu_1)^2}{2\sigma^2}} &= \frac{\pi_2}{\sqrt{2\pi}\sigma} e^{-\frac{(x-\mu_2)^2}{2\sigma^2}}\\ \vdots \end{align}\]
Canceling the common terms, and taking the logarithm of both sides:
\[ \log(\pi_1) - \frac{(x - \mu_1)^2}{2 \sigma^2} = \log(\pi_2) - \frac{(x - \mu_2)^2}{2 \sigma^2} \]
Expanding the quadratic terms:
\[ \log(\pi_1) - \frac{x^2 - 2x \mu_1 + \mu_1^2}{2 \sigma^2} = \log(\pi_2) - \frac{x^2 - 2x \mu_2 + \mu_2^2}{2 \sigma^2} \]
Simplifying both sides:
\[ \log(\pi_1) + \frac{2x \mu_1 - \mu_1^2}{2 \sigma^2} = \log(\pi_2) + \frac{2x \mu_2 - \mu_2^2}{2 \sigma^2} \]
If the left-hand side (LHS) is greater than the right-hand side (RHS), the observation is classified as \(y = 1\). If LHS \(<\) RHS, the observation is classified as \(y = 2\).
We can simplify the expression for each class \(k\) as:
\[ \delta_k(x) = x \frac{\mu_k}{\sigma^2} - \frac{\mu_k^2}{2 \sigma^2} + \log(\pi_k) \]
Discriminant score
This is boils down to to assigning the observation to the class for which discriminant score \(\delta_k(x)\) is the largest: \[\begin{equation} \delta_k(x) = x \cdot \frac{\mu_k}{\sigma^2} - \frac{\mu_{k}^2}{\sigma^2} + \log(\pi_k) \end{equation}\]
Notice the discriminant score is a linear function of \(\boldsymbol x\).
Linear Decision boundary
Case 1: Equal variance \(\sigma_1 = \sigma_2\) (equal priors)
\[\begin{equation} x = \frac{\mu_1^2 - \mu_2^2}{2(\mu_1 - \mu_2)} = \frac{\mu_1 + \mu_2}{2} \end{equation}\]
Proof case 1
\section*{Linear Decision Boundary (Equal Variance, Equal Priors)}
We start by using the for each class. For class 1, the discriminant score is:
\[ \delta_1(x) = x \frac{\mu_1}{\sigma^2} - \frac{\mu_1^2}{2 \sigma^2} + \log(\pi_1) \]
For class 2, the discriminant score is:
\[ \delta_2(x) = x \frac{\mu_2}{\sigma^2} - \frac{\mu_2^2}{2 \sigma^2} + \log(\pi_2) \]
Since we are assuming (\(\pi_1 = \pi_2\)), the prior terms \(\log(\pi_1)\) and \(\log(\pi_2)\) cancel out. Thus, the decision boundary is found by setting the two discriminant scores equal:
\[ \delta_1(x) = \delta_2(x) \]
Substitute the discriminant score equations for \(\delta_1(x)\) and \(\delta_2(x)\):
\[ x \frac{\mu_1}{\sigma^2} - \frac{\mu_1^2}{2 \sigma^2} = x \frac{\mu_2}{\sigma^2} - \frac{\mu_2^2}{2 \sigma^2} \]
Now, group the terms involving \(x\) on one side:
\[ x \left( \frac{\mu_1}{\sigma^2} - \frac{\mu_2}{\sigma^2} \right) = \frac{\mu_1^2}{2 \sigma^2} - \frac{\mu_2^2}{2 \sigma^2} \]
Factor out \(\frac{1}{\sigma^2}\):
\[ x \frac{\mu_1 - \mu_2}{\sigma^2} = \frac{\mu_1^2 - \mu_2^2}{2 \sigma^2} \]
Multiply both sides by \(\sigma^2\) to simplify:
\[ x (\mu_1 - \mu_2) = \frac{\mu_1^2 - \mu_2^2}{2} \]
Finally, solve for \(x\):
\[ x = \frac{\mu_1^2 - \mu_2^2}{2 (\mu_1 - \mu_2)} \]
This simplifies to:
\[ x = \frac{\mu_1 + \mu_2}{2} \]
Thus, the decision boundary is the when the variances are equal and the priors are equal.
Linear Decision boundary
Case 2: Equal variance \(\sigma_1 = \sigma_2\) (unequal priors)
\[\begin{equation} x = \frac{\log \left( \frac{\pi_2}{\pi_1} \right) + \frac{\mu_2^2}{2\sigma^2} - \frac{\mu_1^2}{2\sigma^2}}{\frac{\mu_1}{\sigma^2} - \frac{\mu_2}{\sigma^2}} \end{equation}\]
Proof case 2
The decision boundary \(x\) is found by setting \(\delta_1(x) = \delta_2(x)\)
\[ \frac{x \mu_1}{\sigma^2} - \frac{\mu_1^2}{2\sigma^2} + \log(\pi_1) = \frac{x \mu_2}{\sigma^2} - \frac{\mu_2^2}{2\sigma^2} + \log(\pi_2) \]
Rearranging the terms to isolate \(x\):
\[ \begin{align*} x \left( \frac{\mu_1}{\sigma^2} - \frac{\mu_2}{\sigma^2} \right) &= \log \left( \frac{\pi_2}{\pi_1} \right) + \frac{\mu_2^2}{2\sigma^2} - \frac{\mu_1^2}{2\sigma^2}\\ \implies x &= \frac{\log \left( \frac{\pi_2}{\pi_1} \right) + \frac{\mu_2^2}{2\sigma^2} - \frac{\mu_1^2}{2\sigma^2}}{\frac{\mu_1}{\sigma^2} - \frac{\mu_2}{\sigma^2}} \end{align*} \]
Substituting the parameter values we get:
\[ \begin{align*} x &= \frac{\log \left( \frac{0.2}{0.8} \right) + \frac{(2)^2}{2\cdot(2)^2} - \frac{-3^2}{2\cdot(2)^2}}{\frac{-3}{(2)^2} - \frac{2}{2^2}} &= 0.61 \end{align*} \]
From \(\delta_k(x)\) to probabilities
From \(\delta_k(x)\) to probabilities, once we have estimates \(\hat{\delta}_k(x)\), we can turn these into estimates for class probabilities:
\[\begin{align} \Pr(Y = k \,|\, X = x) = \frac{e^{\hat{\delta}_k(x)}}{\sum_{l=1}^{K} e^{\hat{\delta}_l(x)}} \end{align}\]
So classifying to the largest \(\hat{\delta}_k(x)\) amounts to classifying to the class for which \(\Pr(Y = k \,|\, X = x)\) is largest.
Probability of each class
Where \(\pi_1\) = 0.8, \(\pi_2\) = 0.2, \(\sigma = \sigma_1 = \sigma_2\) = 2, \(\mu_1\) = -3 \(\mu_2\) = 2 and x = 0.61
\[ \begin{align} P(&Y = 1 \mid x = 0.61) \\ &= \frac{\pi_1 f_1(x)}{\pi_1 f_1(x) + \pi_2 f_2(x)}\\ &= \frac{0.8 \cdot 0.04} {0.8 \cdot 0.04 + 0.2 \cdot 0.16}\\ &= 0.5 \end{align} \]
But we don’t know \(f_k\)
No problem. We estimate them!

Parameter Estimates
\[\begin{align} \hat{\pi}_k &= \frac{n_k}{n} \quad\quad \hat{\mu}_k = \frac{1}{n_k} \sum_{i: y_i=k} x_i\\ \hat{\sigma}^2 &= \frac{1}{n - K} \sum_{k=1}^{K} \sum_{i: y_i=k} (x_i - \hat{\mu}_k)^2 \end{align}\]
where \(n\) is the total number of training observations, \(n_k\) is the number of observations in class \(k\), and \(\mu_k\) and \(\hat{\sigma}^2_k\) are the MLE estimates for mean and (pooled) variance in the \(k\)th class.
Classifier
When we do not have equal variance (i.e \(\sigma_1 \neq \sigma_2\)) our classifier predicts \(x_0\) to group 1 if
\[ \pi_1 \cdot f_1(x; \mu_1, \sigma_1) > \pi_2\cdot f_2(x; \mu_2, \sigma_2) \]
More generally, we classify observation \(x\) based on the following rule:
\[ \hat y = \underset{k}{\operatorname{argmax}} \pi_k \cdot f_k(x; \mu_k, \sigma_k) \]
Quadratic Decision boundary
Case 3: Unequal variance \(\sigma_1 \neq \sigma_2\) (unequal priors)
The decision boundary is a quadratic equation in \(x\):
\[ Ax^2 + Bx + C = 0 \]
where:
- \(A = \frac{1}{2\sigma_2^2} - \frac{1}{2\sigma_1^2}\) (coefficient for the quadratic term),
- \(B = \frac{\mu_2}{\sigma_2^2} - \frac{\mu_1}{\sigma_1^2}\) (coefficient for the linear term),
- \(C = \frac{\mu_1^2}{2\sigma_1^2} - \frac{\mu_2^2}{2\sigma_2^2} + \ln\left(\frac{\pi_1 \sigma_2}{\pi_2 \sigma_1}\right)\) (constant term).
Proof for Case 3
\[ \pi_1 f_1(x) = \pi_2 f_2(x) \]
Substitute the normal probability density functions for \(f_1(x)\) and \(f_2(x)\):
\[ \pi_1 \frac{1}{\sqrt{2\pi \sigma_1^2}} \exp\left(-\frac{(x - \mu_1)^2}{2\sigma_1^2}\right) = \pi_2 \frac{1}{\sqrt{2\pi \sigma_2^2}} \exp\left(-\frac{(x - \mu_2)^2}{2\sigma_2^2}\right) \]
Simplifying the constants:
\[ \frac{\pi_1}{\sigma_1} \exp\left(-\frac{(x - \mu_1)^2}{2\sigma_1^2}\right) = \frac{\pi_2}{\sigma_2} \exp\left(-\frac{(x - \mu_2)^2}{2\sigma_2^2}\right) \]
Take the natural logarithm of both sides to remove the exponential terms:
\[ \ln\left(\frac{\pi_1}{\sigma_1}\right) - \frac{(x - \mu_1)^2}{2\sigma_1^2} = \ln\left(\frac{\pi_2}{\sigma_2}\right) - \frac{(x - \mu_2)^2}{2\sigma_2^2} \]
Now expand and rearrange terms:
\[ -\frac{(x - \mu_1)^2}{2\sigma_1^2} + \frac{(x - \mu_2)^2}{2\sigma_2^2} = \ln\left(\frac{\pi_1 \sigma_2}{\pi_2 \sigma_1}\right) \]
Expand the quadratic terms:
\[ -\frac{x^2 - 2x\mu_1 + \mu_1^2}{2\sigma_1^2} + \frac{x^2 - 2x\mu_2 + \mu_2^2}{2\sigma_2^2} = \ln\left(\frac{\pi_1 \sigma_2}{\pi_2 \sigma_1}\right) \]
Simplify the terms involving \(x\):
\[ \left(\frac{1}{2\sigma_2^2} - \frac{1}{2\sigma_1^2}\right) x^2 + \left(\frac{\mu_2}{\sigma_2^2} - \frac{\mu_1}{\sigma_1^2}\right) x + \left(\frac{\mu_1^2}{2\sigma_1^2} - \frac{\mu_2^2}{2\sigma_2^2}\right) = \ln\left(\frac{\pi_1 \sigma_2}{\pi_2 \sigma_1}\right) \]

ESL Figure 4.4: Left: shows some data from three classes, with linear decision boundaries found by LDA. Right: shows quadratic decision boundaries found using QDA.
LDA vs Logistic
Pros of LDA
- If \(n\) is small and the distribution of the predictors \(X\) is approximately normal in each class, the linear discriminant model is more stable than the logistic regression model.
- LDA is popular when we have more than two response classes
Cons of LDA
Assumes normality, i.e. that the features are normally distributed (which is not always appropriate).
Assumes homoscedasticity, meaning the classes share the same covariance matrix (strong assumption)
Multivariate Normal Distribution
For Discriminant analysis when \(p>1\) we use the Multivariate Normal Distribution (MVN).
A MVN random vector \(\mathbf X\), has probability distribution given by \[f(\mathbf x) = \frac{1}{(2\pi)^{p/2}|\boldsymbol{\Sigma}|^{1/2}} e^{-\frac{1}{2}(\mathbf{x}-\boldsymbol{\mu})^T\boldsymbol{\Sigma}^{-1}(\mathbf{x}-\boldsymbol{\mu})}\] with mean vector \(\boldsymbol{\mu}\) and covariance matrix \(\boldsymbol{\Sigma}\).
3D Plot of Multivariate Normal

Covariance Matrix
The covariance matrix is symmetric a square matrix where each entry \(\Sigma_{ij}\) represents the covariance between variables \(X_i\) and \(X_j\)
The diagonal elements (\(\sigma_{i}\)) represent the variances of individual variables, and the off-diagonal elements (\(\sigma_{ij}\) for \(i\neq j\)) represent the covariances between pairs of variables. \[\begin{equation} \boldsymbol \Sigma = \begin{bmatrix} \sigma_{1}^2 & \sigma_{12} & \ldots & \sigma_{1p} \\ \sigma_{12} & \sigma_{2}^2 & \ldots & \sigma_{2p} \\ \vdots & \vdots & \ddots & \vdots \\ \sigma_{1p} & \sigma_{2p} & \ldots & \sigma_{p}^2 \end{bmatrix} \quad \text{ where }\sigma_{ij} = \sigma_{ji} = \text{Cov}(X_i, X_j) \end{equation}\]
Uncorrelated
Code
# Install and load the required libraries
library(plotly)
library(mvtnorm)
# Parameters for the multivariate normal distribution
mean_vec <- c(0, 0) # Mean vector
cov_mat <- matrix(c(1, 0, 0, 1), nrow = 2) # Covariance matrix
# Generate data grid
x <- seq(-3, 3, length.out = 100)
y <- seq(-3, 3, length.out = 100)
grid <- expand.grid(x, y)
# Calculate the multivariate normal density for each point in the grid
z <- dmvnorm(grid, mean = mean_vec, sigma = cov_mat)
# Reshape the z values to match the grid dimensions
z_matrix <- matrix(z, nrow = length(x), ncol = length(y))
# Create an interactive 3D surface plot using plotly
plot_ly(z = ~z_matrix, x = ~x, y = ~y, type = "surface") %>%
layout(scene = list(
xaxis = list(title = "X-axis"),
yaxis = list(title = "Y-axis"),
zaxis = list(title = "Density")
))Code
# Install and load the required libraries
library(plotly)
library(mvtnorm)
# Parameters for the multivariate normal distribution
mean_vec <- c(0, 0) # Mean vector
cov_mat <- matrix(c(1, 0.5, 0.5, 1), nrow = 2) # Covariance matrix
# Generate data grid
x <- seq(-3, 3, length.out = 100)
y <- seq(-3, 3, length.out = 100)
grid <- expand.grid(x, y)
# Calculate the multivariate normal density for each point in the grid
z <- dmvnorm(grid, mean = mean_vec, sigma = cov_mat)
# Reshape the z values to match the grid dimensions
z_matrix <- matrix(z, nrow = length(x), ncol = length(y))
# Create an interactive 3D surface plot using plotly
plot_ly(z = ~z_matrix, x = ~x, y = ~y, type = "surface") %>%
layout(scene = list(
xaxis = list(title = "X-axis"),
yaxis = list(title = "Y-axis"),
zaxis = list(title = "Density")
))LDA when \(p>1\)
In the case of \(p > 1\) predictors, the LDA classifier assumes that the observations in the \(k\)th class are drawn from a multivariate Gaussian distribution \(\mathcal{N}(\boldsymbol\mu_k, \boldsymbol\Sigma)\), where \(\boldsymbol\mu_k\) is a class-specific mean vector, and \(\boldsymbol\Sigma\) is a covariance matrix that is common to all \(K\) classes.
Algebra reveals that the Bayes classifier assigns an observation \(X = x\) to the class for which \[\begin{equation} \delta_k(x) = x^T \boldsymbol\Sigma^{-1} \boldsymbol\mu_k - \frac{1}{2} \boldsymbol\mu_k^T \boldsymbol\Sigma^{-1} \boldsymbol\mu_k + \log(\pi_k) \end{equation}\]
What does equal Covariance look like?
Source of image [1]
Contour Plots (equal covariance)
Code
par(mar=c(5,4,1.1,0.1))
# Define the means and shared covariance matrix for the two classes
mean1 <- c(0, 0)
mean2 <- c(3, 3)
cov_matrix <- matrix(c(1, 0.5, 0.5, 1), ncol=2) # Same covariance matrix for both classes
# Create a grid of points
x <- seq(-3, 6, length=100)
y <- seq(-3, 6, length=100)
grid <- expand.grid(X1 = x, X2 = y)
# Calculate the multivariate normal densities for each class
z1 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean1, sigma=cov_matrix))
z2 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean2, sigma=cov_matrix))
# Reshape for contour plotting
z1_matrix <- matrix(z1, nrow=length(x), ncol=length(y))
z2_matrix <- matrix(z2, nrow=length(x), ncol=length(y))
# Plot the contour plots for the two classes
contour(x, y, z1_matrix, drawlabels=FALSE, col="blue", lwd=2, main="Equal Covariance", xlab="X1", ylab="X2")
contour(x, y, z2_matrix, drawlabels=FALSE, col="red", lwd=2, add=TRUE)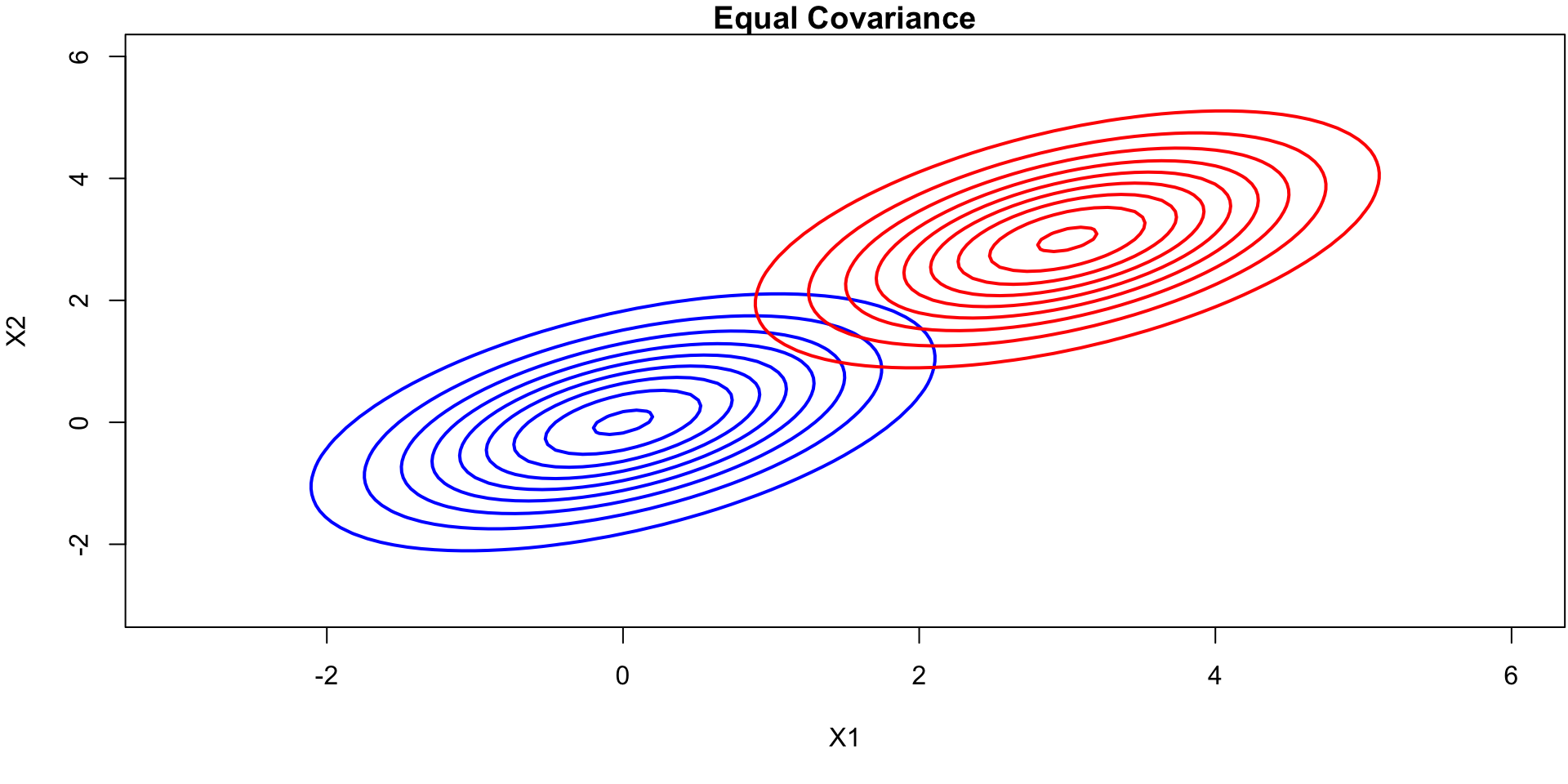
Contour Plots (unequal covariance)
Code
# Load necessary libraries
library(mvtnorm)
par(mar=c(5,4,1.1,0.1))
# Define the means and covariance matrices for the two classes
mean1 <- c(0, 0)
cov1 <- matrix(c(1, 0.25, 0.25, 1), ncol=2)
mean2 <- c(3, 3)
cov2 <- matrix(c(1, -0.75, -0.75, 1), ncol=2)
# Create a grid of points
x <- seq(-3, 6, length=100)
y <- seq(-3, 6, length=100)
grid <- expand.grid(X1 = x, X2 = y)
# Calculate the multivariate normal densities for each class
z1 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean1, sigma=cov1))
z2 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean2, sigma=cov2))
# Reshape for contour plotting
z1_matrix <- matrix(z1, nrow=length(x), ncol=length(y))
z2_matrix <- matrix(z2, nrow=length(x), ncol=length(y))
# Plot the contour plots for the two classes
contour(x, y, z1_matrix, drawlabels=FALSE, col="blue", lwd=2, main="Unequal Covariance", xlab="X1", ylab="X2")
contour(x, y, z2_matrix, drawlabels=FALSE, col="red", lwd=2, add=TRUE)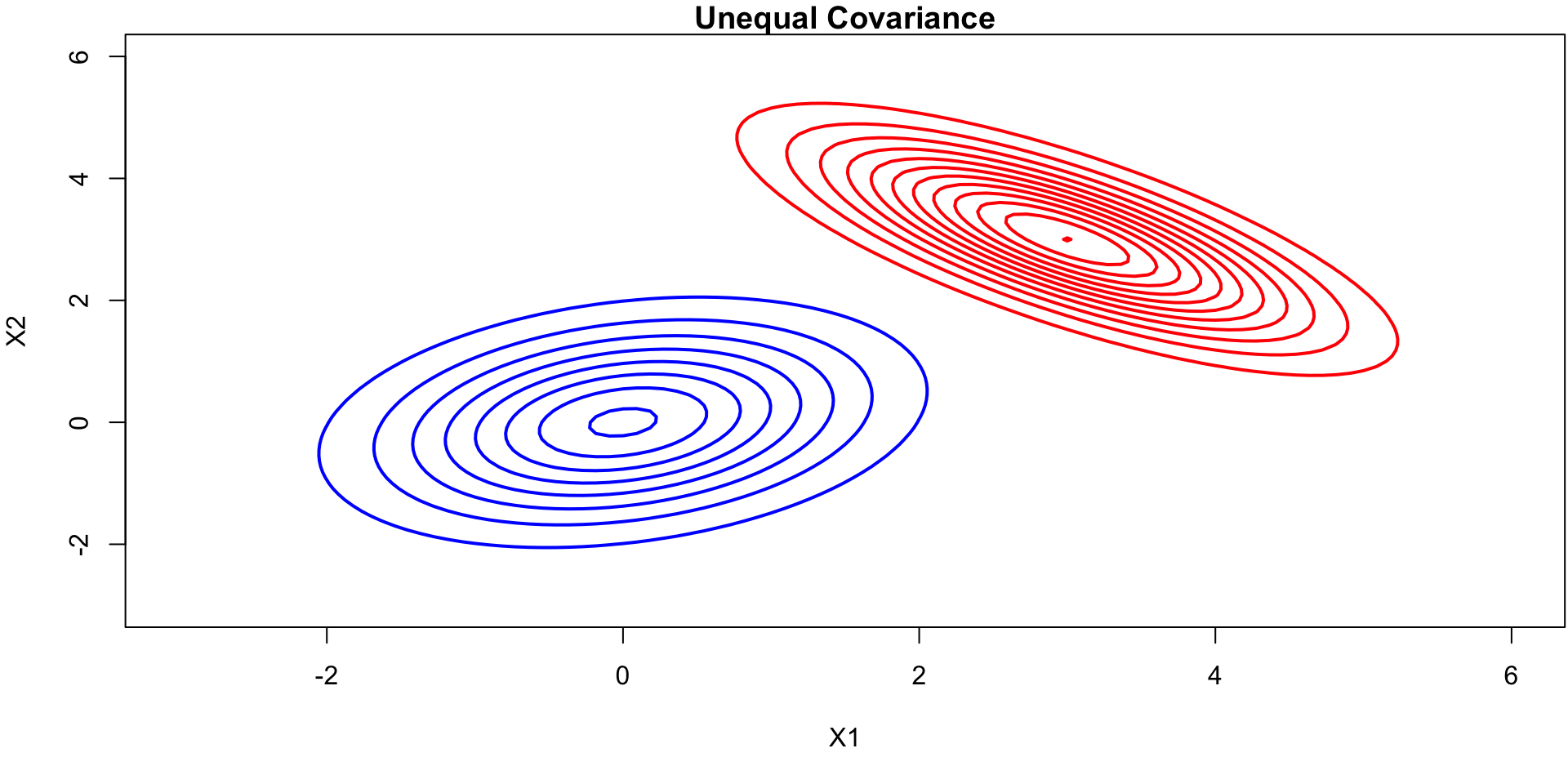
LDA QDA in 2D
Code
## | cache: true
# Load necessary libraries
library(MASS)
par(mar=c(5,4,1.1,0.1))
# Define the means and shared covariance matrix for the two classes
mean1 <- c(0, 0)
mean2 <- c(3, 3)
cov_matrix <- matrix(c(1, 0.5, 0.5, 1), ncol=2) # Same covariance matrix for both classes
# Create a grid of points
x <- seq(-3, 6, length=100)
y <- seq(-3, 6, length=100)
grid <- expand.grid(X1 = x, X2 = y)
# Calculate the multivariate normal densities for each class
z1 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean1, sigma=cov_matrix))
z2 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean2, sigma=cov_matrix))
# Reshape for contour plotting
z1_matrix <- matrix(z1, nrow=length(x), ncol=length(y))
z2_matrix <- matrix(z2, nrow=length(x), ncol=length(y))
# Plot the contour plots for the two classes
contour(x, y, z1_matrix, drawlabels=FALSE, col="blue", lwd=2, main="LDA Decision Boundary", xlab="X1", ylab="X2")
contour(x, y, z2_matrix, drawlabels=FALSE, col="red", lwd=2, add=TRUE)
# Plot the LDA decision boundary (it will be linear)
contour(x, y, z1_matrix - z2_matrix, levels=0, drawlabels=FALSE, col="green", lwd=2, lty=2, add=TRUE)
# Add a title and labels
title(main="LDA Decision Boundary")
Code
##| cache: true
# Load necessary libraries
library(mvtnorm)
par(mar=c(5,4,1.1,0.1))
# Define the means and covariance matrices for the two classes
mean1 <- c(0, 0)
cov1 <- matrix(c(1, 0.5, 0.5, 1), ncol=2)
mean2 <- c(3, 3)
cov2 <- matrix(c(1, -0.5, -0.5, 1), ncol=2)
# Create a grid of points
x <- seq(-3, 6, length=100)
y <- seq(-3, 6, length=100)
grid <- expand.grid(X1 = x, X2 = y)
# Calculate the multivariate normal densities for each class
z1 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean1, sigma=cov1))
z2 <- apply(grid, 1, function(x) dmvnorm(x, mean=mean2, sigma=cov2))
# Reshape for contour plotting
z1_matrix <- matrix(z1, nrow=length(x), ncol=length(y))
z2_matrix <- matrix(z2, nrow=length(x), ncol=length(y))
# Plot the contour plots for the two classes
contour(x, y, z1_matrix, drawlabels=FALSE, col="blue", lwd=2, main="QDA Decision Boundary", xlab="X1", ylab="X2")
contour(x, y, z2_matrix, drawlabels=FALSE, col="red", lwd=2, add=TRUE)
# Plot the QDA decision boundary where the two densities are equal
contour(x, y, z1_matrix - z2_matrix, levels=0, drawlabels=FALSE, col="green", lwd=2, lty=2, add=TRUE)
Illustration \(p = 2\) and \(K=3\)

ILSR Fig 4.6: Here \(\pi_1 = \pi_2 = \pi_3\) Left: Ellipses that contain 95 % of the probability for each of the three classes are shown. The dashed lines are the Bayes decision boundaries. Right: 20 observations were generated from each class, and the corresponding LDA decision boundaries are indicated using solid black lines. The Bayes decision boundaries are shown as dashed lines.
LDA to QDA
When \(f_k(x)\) are Gaussian densities, with the same covariance matrix \(\boldsymbol\Sigma\) in each class, this leads to linear discriminant analysis.
With Gaussians but different \(\boldsymbol \Sigma_k\) in each class, we get quadratic discriminant analysis.
Because the \(\boldsymbol\Sigma_k\) are different, we don’t loose the quadratic terms so we get discriminant function is now
\[\begin{equation} \delta_k(x) = -\frac{1}{2}(x - \mu_k)^T \boldsymbol \Sigma_k^{-1} (x - \mu_k) + \log \pi_k - \frac{1}{2} \log |\Sigma_k| \end{equation}\]
QDA Parameter Estimates
For QDA/LDA the parameters estimates are given by: \[ \begin{align} \hat{\mu}_k &= \frac{1}{n_k} \sum_{i : y_i = k} x_i, & \hat{\pi}_k &= \frac{n_k}{n} \end{align} \]
For QDA the estimate for covariance is: \[\hat{\Sigma}_k = \frac{1}{n_k - 1} \sum_{i : y_i = k} (x_i - \mu_k)(x_i - \mu_k)^\top,\]
LDA Parameter Estimates
For QDA/LDA the parameters estimates are given by: \[ \begin{align} \hat{\mu}_k &= \frac{1}{n_k} \sum_{i : y_i = k} x_i, & \hat{\pi}_k &= \frac{n_k}{n} \end{align} \]
For LDA, one uses the pooled covariance estimator:
\[\hat{\Sigma} = \frac{1}{n - K} \sum_{k=1}^{K} \sum_{i : y_i = k} (x_i - \mu_k)(x_i - \mu_k)^\top.\]
Summary
Both LDA and QDA assumes each class follows an underlying Gaussian distribution
LDA
assumes equal variance across classes
PRO: easier to fit
CON: less flexible
QDA
doesn’t assume equal variance across classes
PRO: often leads to a lower error rate
CON: needs more data
Bias-variance tradeoff: the LDA can be too simple whereas the QDA has a higher risk of overfitting to our data.
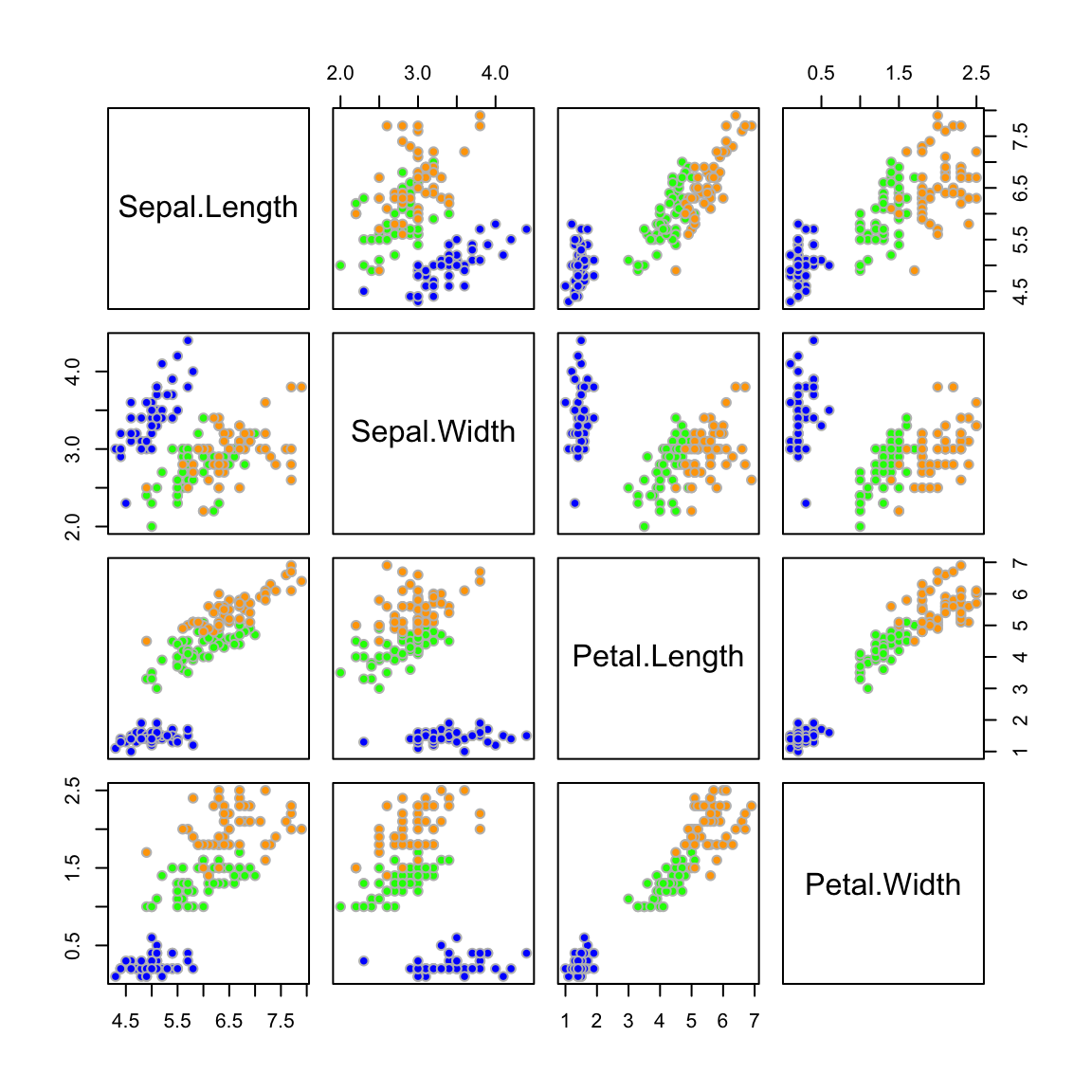
Example: Iris
LDA fitted model
Let’s fit the LDA classifier using all 4 predictors
...
Prior probabilities of groups:
setosa versicolor virginica
0.3333333 0.3333333 0.3333333
Group means:
Sepal.Length Sepal.Width Petal.Length Petal.Width
setosa 5.006 3.428 1.462 0.246
versicolor 5.936 2.770 4.260 1.326
virginica 6.588 2.974 5.552 2.026
...Plotted fit
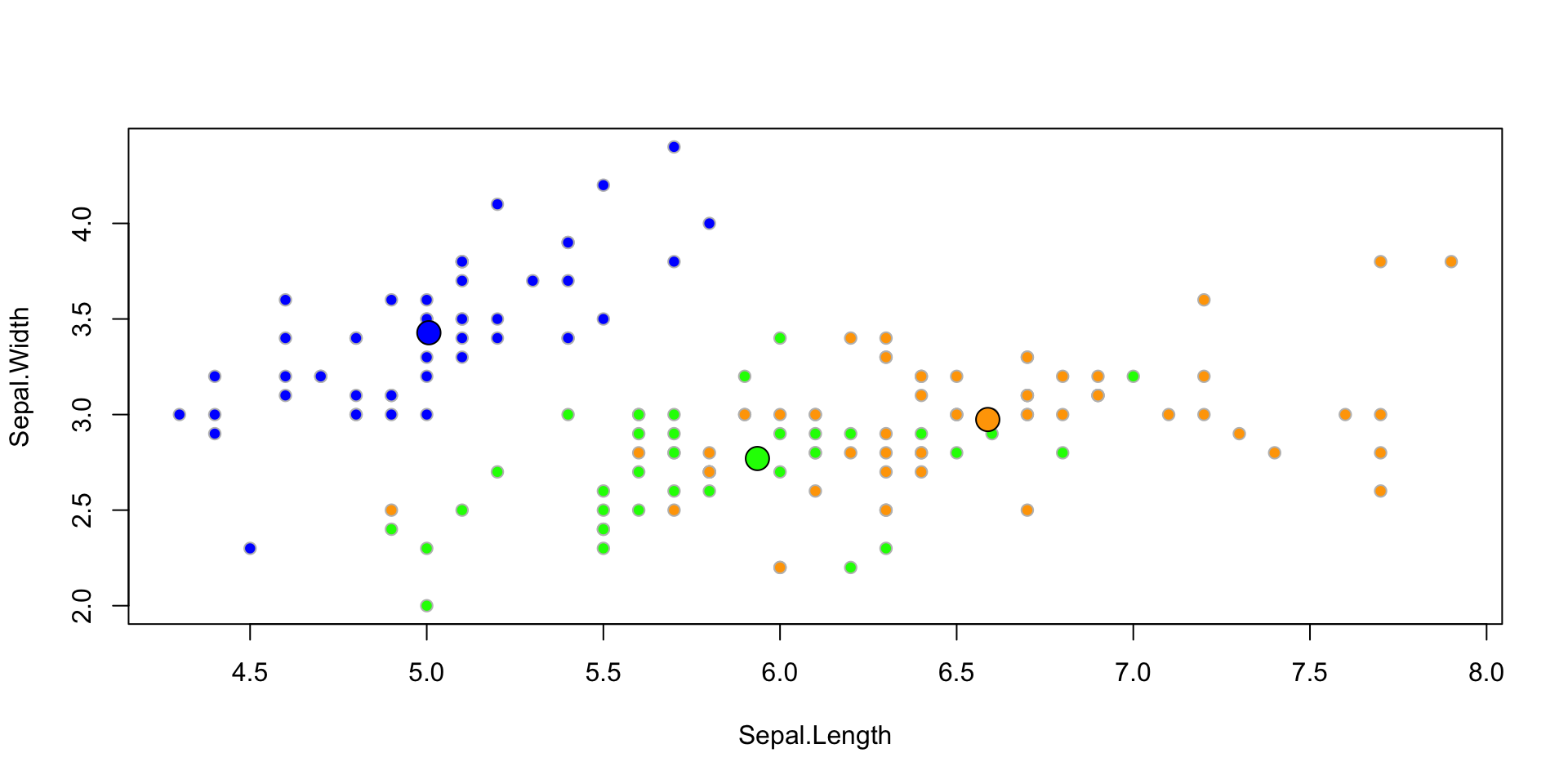
Classification Table
LDA classifies all but 3 of the 150 training samples correctly.
[1] "class" "posterior" "x"
setosa versicolor virginica
setosa 50 0 0
versicolor 0 48 2
virginica 0 1 49This of course is the training error so it may be overly optimistic.
Posterior Probabilties
We can obtain the posterior probabilities if we so desire:
setosa versicolor virginica
1 1 0.000 0.000
2 1 0.000 0.000
3 1 0.000 0.000
4 1 0.000 0.000
5 1 0.000 0.000
6 1 0.000 0.000
7 1 0.000 0.000
8 1 0.000 0.000
9 1 0.000 0.000
10 1 0.000 0.000
11 1 0.000 0.000
12 1 0.000 0.000
13 1 0.000 0.000
14 1 0.000 0.000
15 1 0.000 0.000
16 1 0.000 0.000
17 1 0.000 0.000
18 1 0.000 0.000
19 1 0.000 0.000
20 1 0.000 0.000
21 1 0.000 0.000
22 1 0.000 0.000
23 1 0.000 0.000
24 1 0.000 0.000
25 1 0.000 0.000
26 1 0.000 0.000
27 1 0.000 0.000
28 1 0.000 0.000
29 1 0.000 0.000
30 1 0.000 0.000
31 1 0.000 0.000
32 1 0.000 0.000
33 1 0.000 0.000
34 1 0.000 0.000
35 1 0.000 0.000
36 1 0.000 0.000
37 1 0.000 0.000
38 1 0.000 0.000
39 1 0.000 0.000
40 1 0.000 0.000
41 1 0.000 0.000
42 1 0.000 0.000
43 1 0.000 0.000
44 1 0.000 0.000
45 1 0.000 0.000
46 1 0.000 0.000
47 1 0.000 0.000
48 1 0.000 0.000
49 1 0.000 0.000
50 1 0.000 0.000
51 0 1.000 0.000
52 0 0.999 0.001
53 0 0.996 0.004
54 0 1.000 0.000
55 0 0.996 0.004
56 0 0.999 0.001
57 0 0.986 0.014
58 0 1.000 0.000
59 0 1.000 0.000
60 0 1.000 0.000
61 0 1.000 0.000
62 0 0.999 0.001
63 0 1.000 0.000
64 0 0.994 0.006
65 0 1.000 0.000
66 0 1.000 0.000
67 0 0.981 0.019
68 0 1.000 0.000
69 0 0.960 0.040
70 0 1.000 0.000
71 0 0.253 0.747
72 0 1.000 0.000
73 0 0.816 0.184
74 0 1.000 0.000
75 0 1.000 0.000
76 0 1.000 0.000
77 0 0.998 0.002
78 0 0.689 0.311
79 0 0.993 0.007
80 0 1.000 0.000
81 0 1.000 0.000
82 0 1.000 0.000
83 0 1.000 0.000
84 0 0.143 0.857
85 0 0.964 0.036
86 0 0.994 0.006
87 0 0.998 0.002
88 0 0.999 0.001
89 0 1.000 0.000
90 0 1.000 0.000
91 0 0.999 0.001
92 0 0.998 0.002
93 0 1.000 0.000
94 0 1.000 0.000
95 0 1.000 0.000
96 0 1.000 0.000
97 0 1.000 0.000
98 0 1.000 0.000
99 0 1.000 0.000
100 0 1.000 0.000
101 0 0.000 1.000
102 0 0.001 0.999
103 0 0.000 1.000
104 0 0.001 0.999
105 0 0.000 1.000
106 0 0.000 1.000
107 0 0.049 0.951
108 0 0.000 1.000
109 0 0.000 1.000
110 0 0.000 1.000
111 0 0.013 0.987
112 0 0.002 0.998
113 0 0.000 1.000
114 0 0.000 1.000
115 0 0.000 1.000
116 0 0.000 1.000
117 0 0.006 0.994
118 0 0.000 1.000
119 0 0.000 1.000
120 0 0.221 0.779
121 0 0.000 1.000
122 0 0.001 0.999
123 0 0.000 1.000
124 0 0.097 0.903
125 0 0.000 1.000
126 0 0.003 0.997
127 0 0.188 0.812
128 0 0.134 0.866
129 0 0.000 1.000
130 0 0.104 0.896
131 0 0.000 1.000
132 0 0.001 0.999
133 0 0.000 1.000
134 0 0.729 0.271
135 0 0.066 0.934
136 0 0.000 1.000
137 0 0.000 1.000
138 0 0.006 0.994
139 0 0.193 0.807
140 0 0.001 0.999
141 0 0.000 1.000
142 0 0.000 1.000
143 0 0.001 0.999
144 0 0.000 1.000
145 0 0.000 1.000
146 0 0.000 1.000
147 0 0.006 0.994
148 0 0.003 0.997
149 0 0.000 1.000
150 0 0.018 0.982Fitting QDA
We would similarly fit QDA to the iris data using the qda() function.
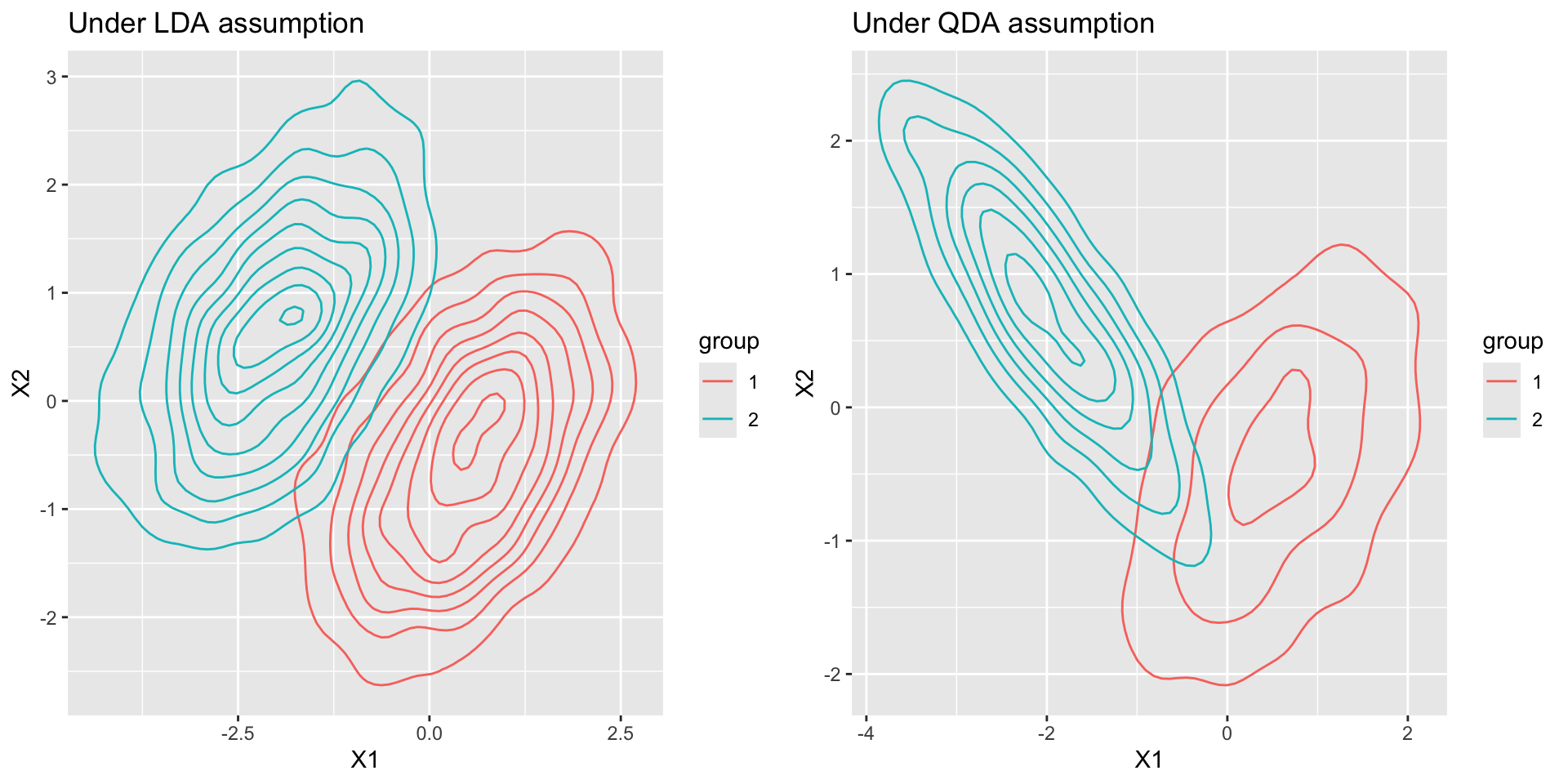
Visualization


iClicker
iClicker
Which of the following is an advantage of QDA over LDA?
It is simpler to implement than LDA
It performs better when the covariance matrices of the classes are equal
It is more flexible and can handle non-linear decision boundaries
It requires fewer data points to estimate parameters
Correct answer: C.
Assumptions
LDA assumes the data is:
LDA makes no assumptions on the data
Gaussian with different covariance for each class
Gaussian with equal covariance for all classes
Gaussian with equal means for call classes
Correct answer: C.