Data 311: Machine Learning
Lecture 15: Boosting
University of British Columbia Okanagan
Recap
Last time we saw how bootstrap aggregation (or bagging) can be used to improve decision trees by reducing its variance.
For Random Forest (RF) we use a similar method to bagging, however, we only consider m predictors at each split1.
That added randomness in RF further reduce the variance of the ML method, thus improving the performance even more.
Introduction
We now discuss boosting, yet another approach for improving the predictions resulting from a decision tree
Again, we focus on decision trees, however, this is a general approach that can be used on many ML methods for regression or classification.
Like bagging and random forests, boosting involves aggregating a large number of decision trees; that is to say, it is another ensemble method.
From bagging to boosting
Some differences moving from bagging to boosting:
- no more bootstrap sampling
- trees are grown sequentially (as opposed to in parallel)
- trees are added together rather than averaged.
- rather than fitting “bushy” trees we prefer “stumps”
Bagging
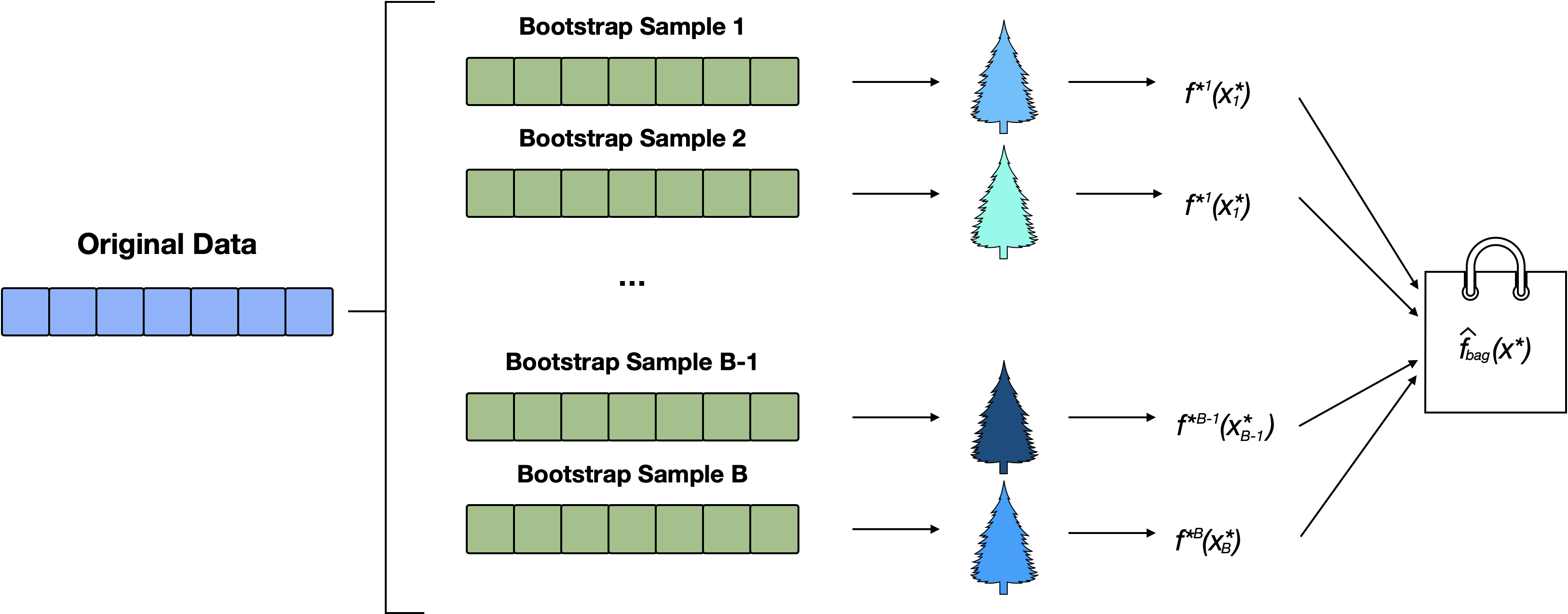
Bagging is an ensemble algorithm that fits multiple multiple trees on different bootstrap samples, then combines the predictions from all models in a single bagged prediction.
Random Forests
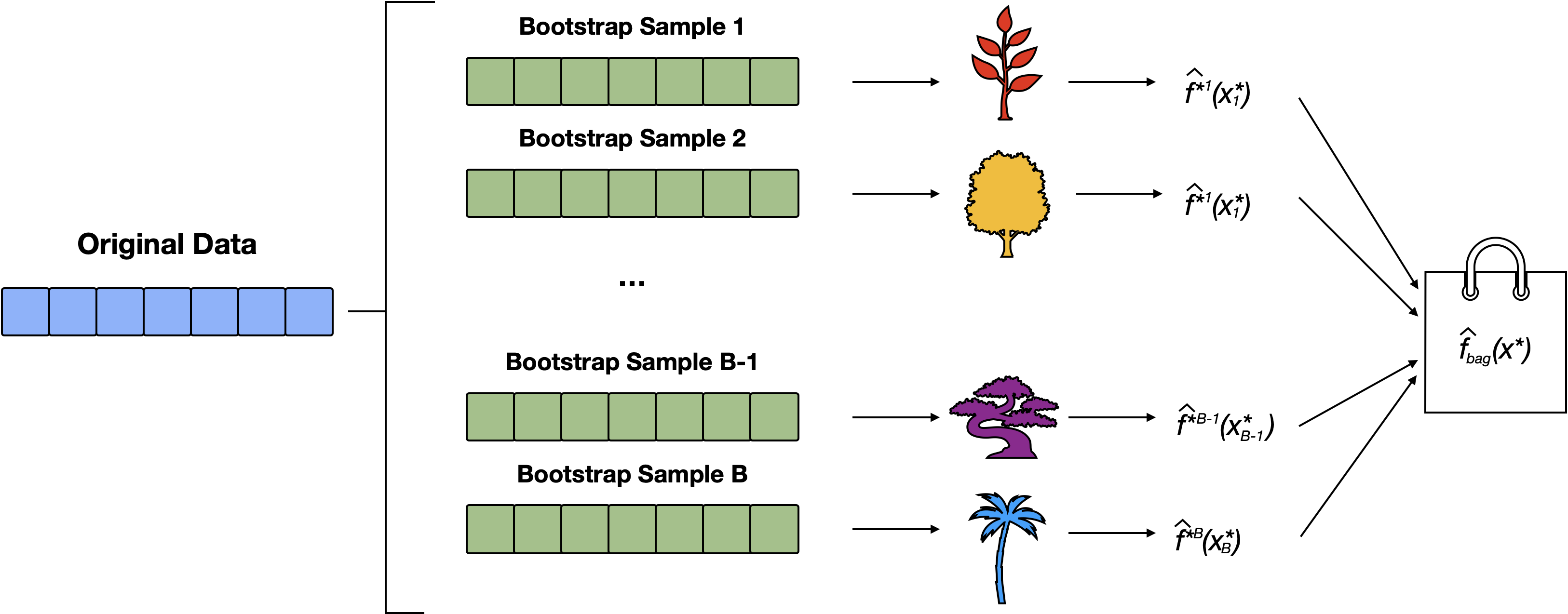
Random forest is an extension of bagging that randomly selects subsets of predictors at each split (thereby diversifying the forest)
From Bushy Tree to Stumps
You may recall that when we make a RF, we fit full sized (aka bushy) trees.
Note that these trees (by design) may look quite different from one another (i.e. they will have difference decision rules, different sizes, etc…)
In boosting, we make a forest of “short” trees (eg. \(d\)1= 1, 4, 8, 32); maybe having one node and two leaves (aka stumps)
These stumps are referred to as a “weak learners”2
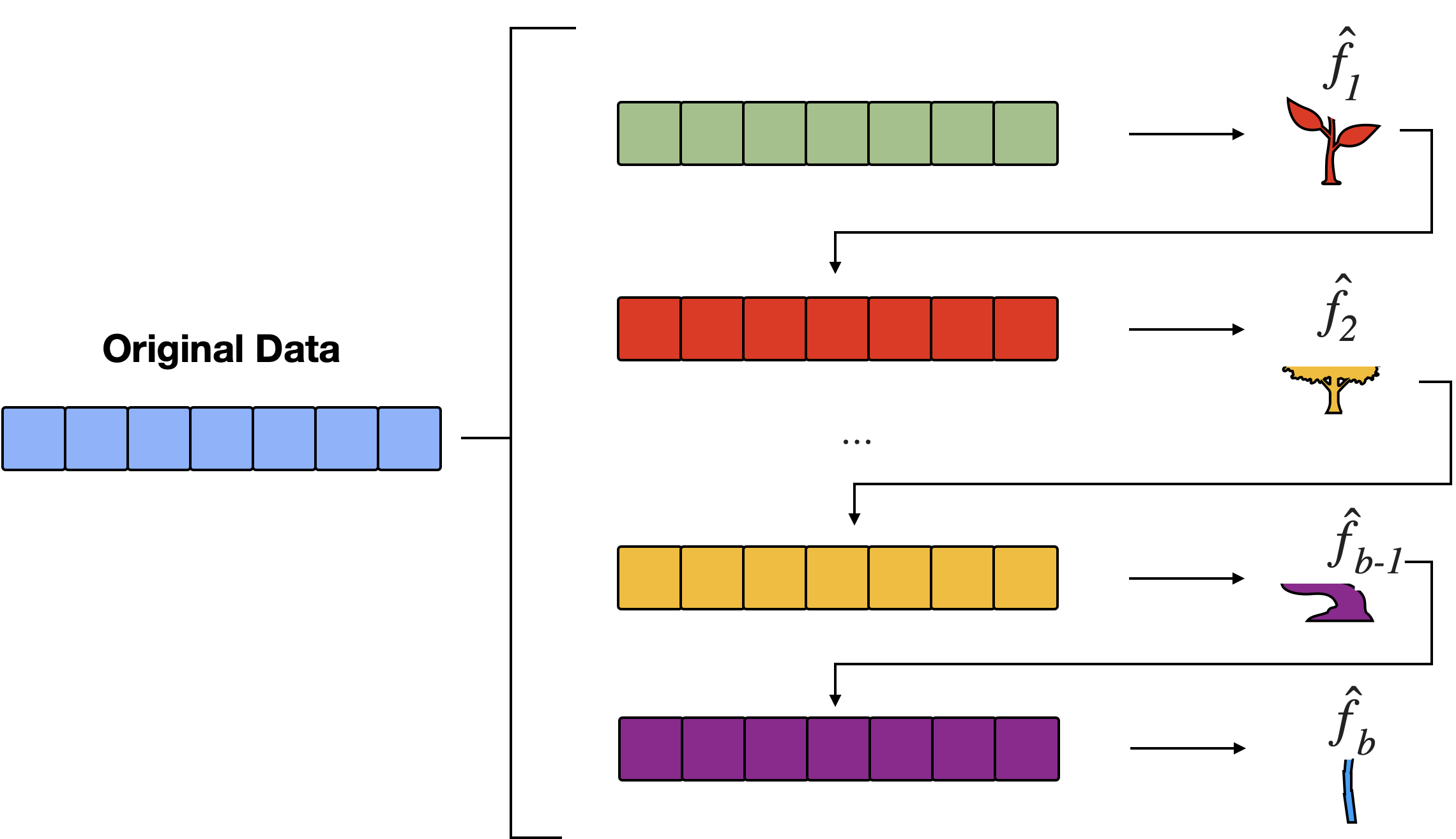
Boosting stumps

Bye-bye boostrap
Boosting does not involve bootstrap sampling; instead each tree is fit to the residuals obtained from the last tree.
That is, we fit a tree using the current residuals, rather than the outcome \(Y\), as the response.
This approach will “attack” large residuals, i.e. will focus on areas where the system is not performing well.
Boosting vs. Bagging/RF
Apart from the boostrap samples, there are other important differences between boosting and random forests:
Trees will be grown sequentially such that each tree will be influenced by the tree grown directly before it.
Like RF, boosting involves combing a larger number of decision trees \(\hat f_1, \dots, \hat f_B\), however…
rather than taking the average (or majority vote) of the corresponding predicted values, the combined prediction is typically a weighted average (or a voting mechanism).
Benefits
Improved Accuracy: By focusing on previously misclassified data points, boosting reduces bias and helps the model fit the training data more accurately.
Reduced Overfitting: Boosting also reduces the risk of overfitting, as it combines the predictions of multiple weak learners, which helps generalize the model.
Robustness: It can handle noisy data and outliers because it adapts to misclassified points.
How to ensemble
By fitting small1 trees to the residuals, we slowly improve \(\hat f\) in areas where it does not perform well.
To slow this down even further, we multiple this fit by a small positive value set by our shrinkage parameter \(\lambda\).
We then add this shrunken decision tree into the fitted function to produce a small incremental nudge in the right direction
In general, statistical learning approaches that “learn slowly” tend to perform well

Algorithm
- Set \(\hat f(x)=0\) and \(r_i =y_i\) for all \(i\) in the data set \((X, r)\).
- For \(b= 1,\dots, B\), repeat:
Fit a tree \(\hat f_b\) with \(d\) splits (\(d+1\) terminal nodes) to \((X,r)\)
Add a shrunken version off this decision tree to your fit: \[\begin{equation} \hat f(x) = \hat f(x) + \lambda \hat f_b(x) \end{equation}\]
Update residuals \(r_i = r_i - \lambda \hat f_b(x)\)
- Output the boosted model \(\hat f(x) = \sum_{b=1}^B \lambda \hat f_b(x)\)
Gene expression Data

Results from performing boosting and random forests on the 15-class gene expression data set in order to predict cancer versus normal. The test error is displayed as a function of the number of trees. For the two boosted models, \(\lambda\) = 0.01. Depth-1 trees slightly outperform depth-2 trees, and both outperform the random forest, although the standard errors are around 0.02, making none of these differences significant. The test error rate for a single tree is 24 %.
Boosting Tuning Parameters
Boosting requires several tuning parameters:
-
\(B\) = the number of fits (trees for today’s example)
- Unlike bagging/RF, boosting can overfit if \(B\) is too large1
-
\(\lambda\) = learning rate for algorithm, a small positive number
- e.g. 0.01 or 0.001 are common
-
\(d\) = complexity or interaction depth (often \(d=1\) works well)
- for trees, this is the number of rules (i.e. internal/decision nodes) so \(d+1\) is the number of terminal nodes.
Example: Regression Simulation
Recall the regression simulation from our CART lecture:

Step 1:
Set \(\hat f(x)=0\) and \(r_i =y_i\) for all \(i\) in the data set \((X, y)\).
Step 2a:
For \(b= 1,\dots, B\), fit a tree \(\hat f_b\) with \(d\) splits (\(d+1\) terminal nodes) to \((X,r)\)
see ?tree.control():
-
mincut= min obs to include in children nodes (default 5) -
minsize= smallest allowed node size (default 10) -
mindev= within-node deviance must be at least this times that of the root node for the node to be split (default = 0.01)
\(\hat f_1(x)\)


Step 2b:
… add a shrunken version off this decision tree to your fit:
Shrunken Tree
\(\hat f(x) = 0 + \lambda \hat f_1(x)\)

Step 2c:
… update residuals \(r_i = r_i - \lambda \ \hat f_b(x)\)
Updated residuals
\(r_i = r_i - \lambda \hat f_1(x)\)

Set \(b = b + 1\), rinse and repeat.
Algorithm Visualization

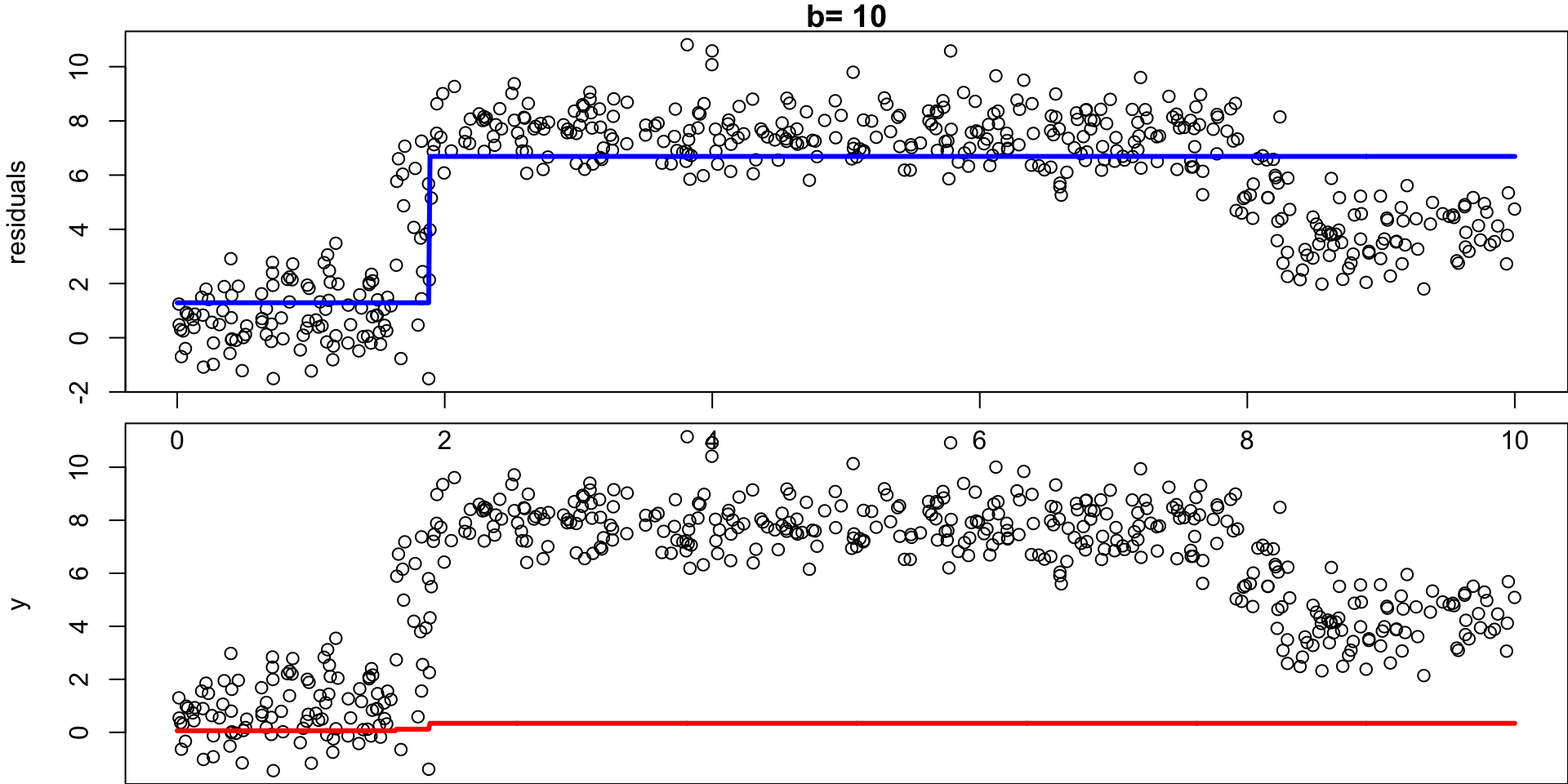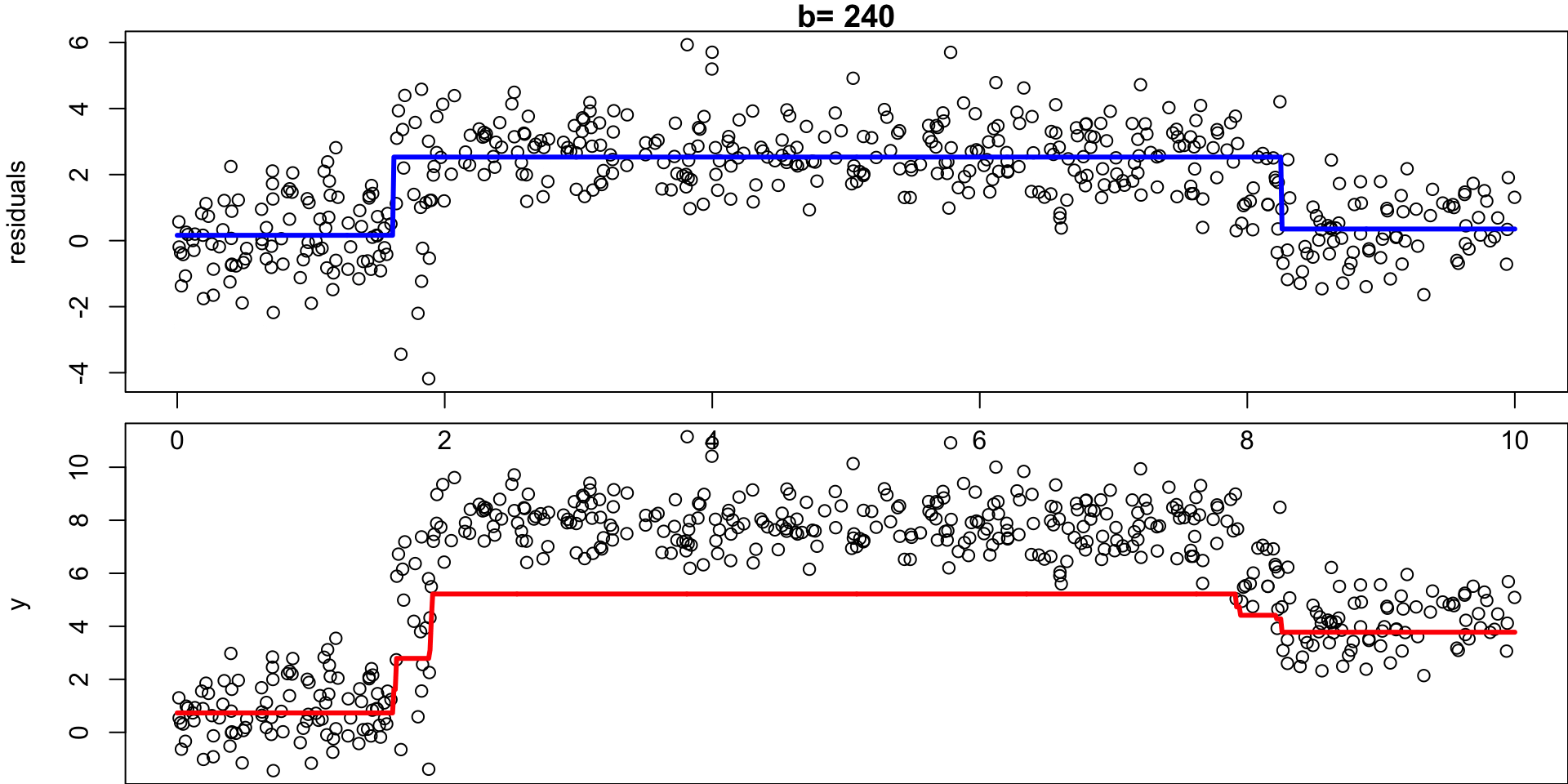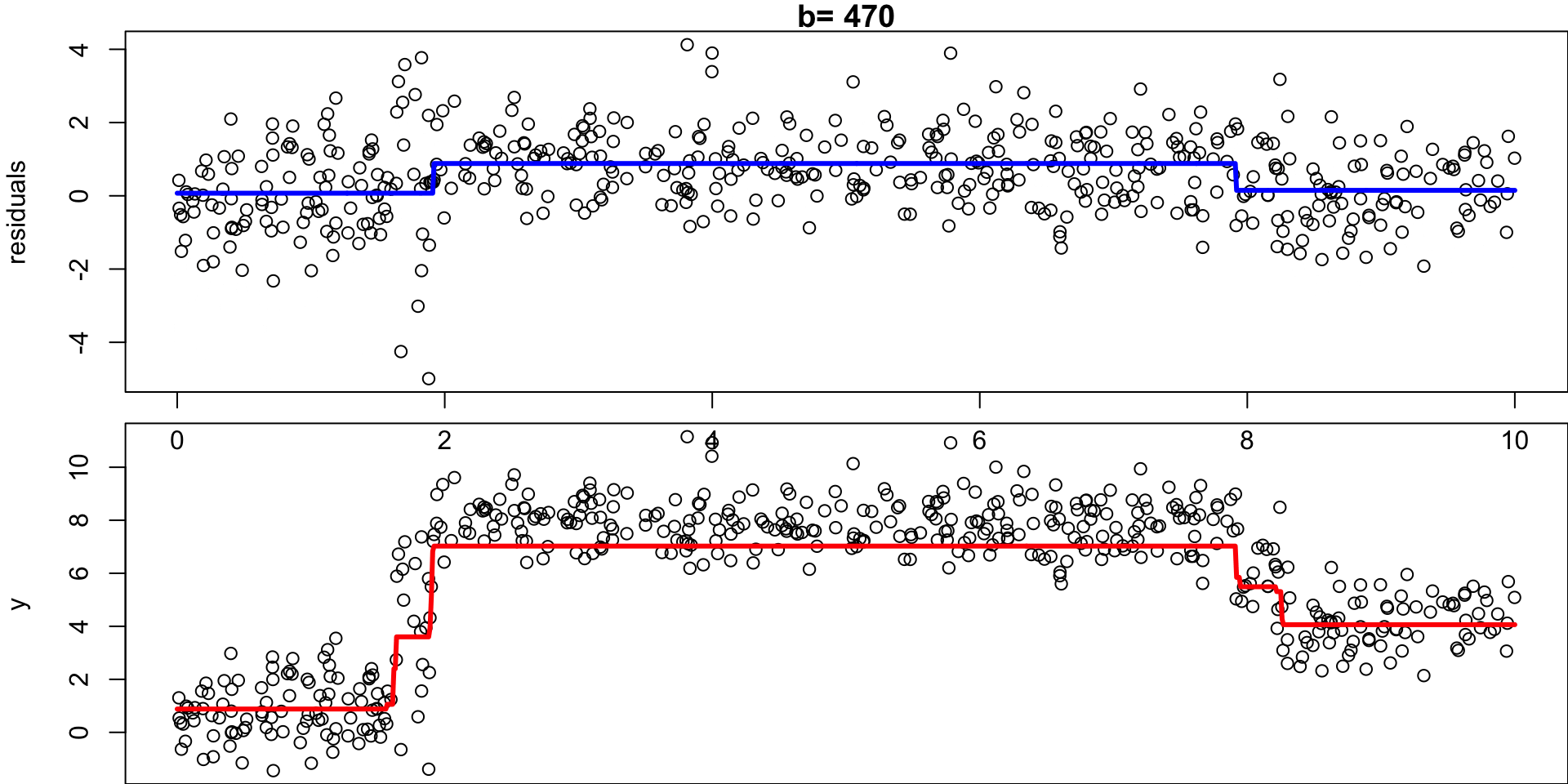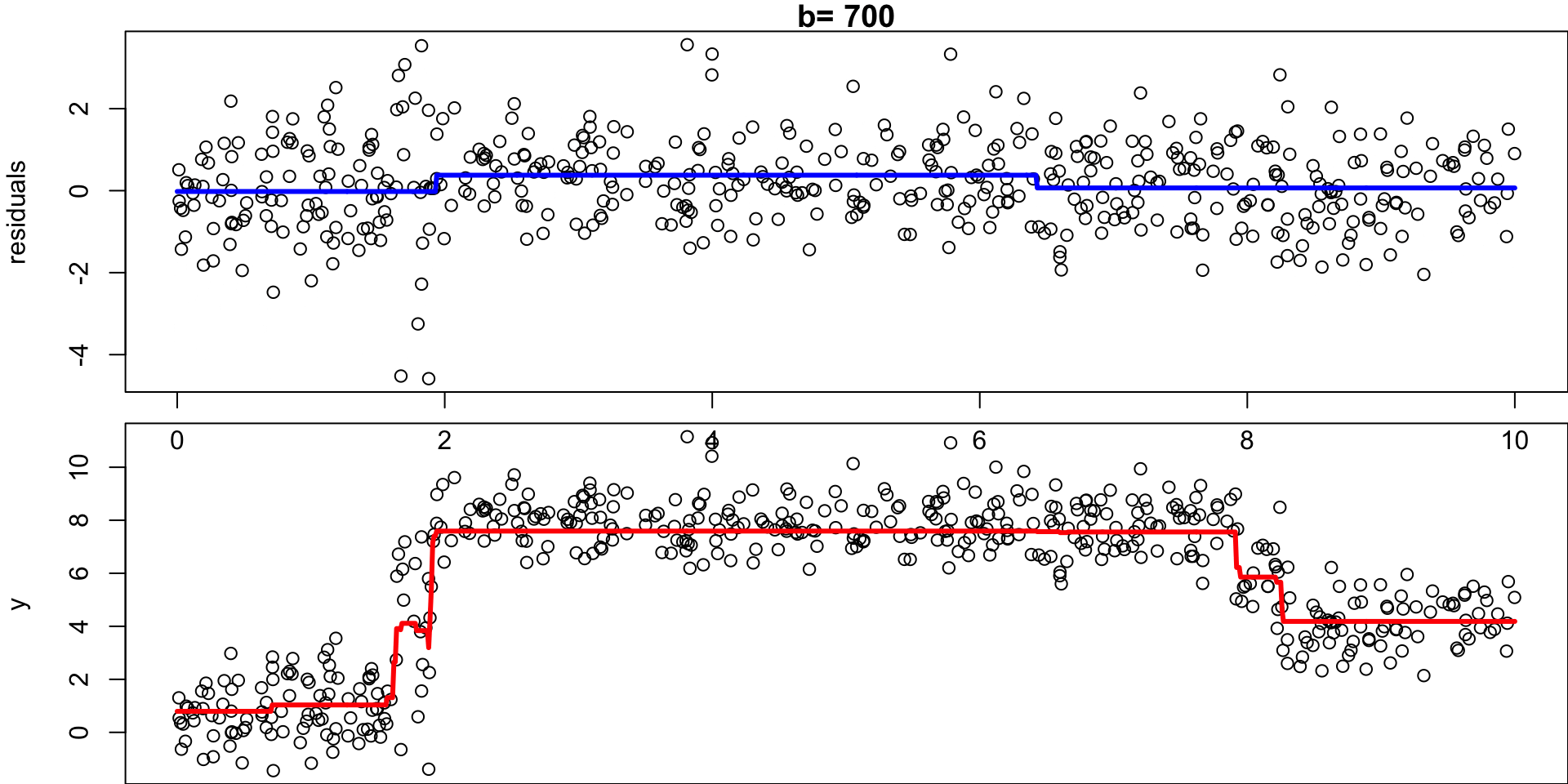
Algorithm Comments
- Given the current model, we fit a decision tree to the residuals (each tree relies on the previous tree)
- We then add a shrunken version of this decision tree to the fitted function \(\hat f\) (to nudge it a bit in the right direction)
- Each tree can be rather small (determined by \(d\))1
By fitting small trees to the residuals, we slowly improve \(\hat f\) in areas where it does not perform well.
Step 3
Output the boosted model \(\hat f(x) = \sum_{b=1}^B \lambda \hat f_b(x)\)

Boosting for classification
Boosting for classification is similar in spirit to boosting for regression, but it is a bit more complex
We will not go into detail here, nor does the ISLR book
Interested students can learn about the details in Elements of Statistical Learning, Chapter 10
To perform boosting with trees (both in the classification and regression setting), we can use the
gbm()(gradient boosted models) function from the gbm package.
Boosting function
-
distribution= “bernoulli” (for binary 0-1 responses), “gaussian” (for regression), “multinomial” for multi-class
Tuning parameters:
-
interaction.depth= \(d\), -
shrinkage= \(\lambda\) -
n.trees= \(B\).
Example: Wine
Let’s perform boosting on the wine data set from the gclus package. As this is a multi-class classification problem, be advised that running this will produce a warning.
Example: Wine
To get a sense of the test error and compare with bagging and RF, we can again use a cross-validation approach (here LOOCV)
library(gbm)
library(gclus); data(wine)
cvboost <- NA
for(i in 1:nrow(wine)){
# preform boosting on the cross-validation training set
weak.lrn <- gbm(factor(Class)~., distribution="multinomial",
data=wine[-i,], n.trees=5000,
interaction.depth=1)
# Prob x_i belongs to the g = 1, 2, 3 group
pig = predict(weak.lrn, n.trees=5000,
newdata=wine[i,], type="response")
# predict the class for the ith validation observation
cvboost[i] <- which.max(pig)
}CV error rate
cvboost
1 2 3
1 58 1 0
2 1 70 0
3 0 0 48This cross validated error rate is 1.12%. In this case boosting is equivalent to RF in terms of prediction. Recall:
Example: body
Recall the body data from the gclus pacakge:
It contains 21 body dimension measurements as well as
age,weight,height, andgenderon 507 individuals;so \(p = 24\) (treating
genderas our binary response)
body: Boosting
set.seed(45613)
library(gbm); data(body) # code doesn't work is Gender is factor
train <- sample(1:nrow(body), round(0.7*nrow(body)))
bodyboost <- gbm(Gender~., distribution="bernoulli", data=body[train,],
n.trees=5000, interaction.depth=1)
pprobs <- predict(bodyboost, newdata=body[-train, ], type="response",
n.trees=5000)
(boosttab <- table(body$Gender[-train], pprobs>0.5 ))
FALSE TRUE
0 74 0
1 5 73Test misclassification rate is 3.29%
body: Random Forests
set.seed(4521)
library(randomForest)
(bodyRF <- randomForest(factor(Gender)~., data=body, importance=TRUE))
Call:
randomForest(formula = factor(Gender) ~ ., data = body, importance = TRUE)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 4
OOB estimate of error rate: 3.94%
Confusion matrix:
0 1 class.error
0 251 9 0.03461538
1 11 236 0.04453441Boosting with cross-validation
# be patient in lab, code will take some time to run
load("data/boostbodyCV.RData")
(tab <- table(body$Gender, as.numeric(cvboost)))
0 1
0 257 3
1 4 243Misclassification rate as estimated by CV is 1.38%
Example: beer
- Consider the
beerdata analyzed in previous lectures.
- Using beer price as our outcome, the cross validated boosting RSS is 97.9 (i.e. its worst of the options we’ve tried)
- Notable the training RSS is 3.86 (code omitted - see lab)
What is this an indication of?
- Ans: that boosting is overfitting to the
beerdata.
Variable Important measure
-
In bagging and RF we saw how we got a measure of Variable Importance which essentially ranks the predictors.
- In regression we record the total amount that the RSS is decreased due to splits over a given predictor, averaged over all \(B\) trees.
- In classification, we add up the total among that the Gini index is decreased by splits over a given predictor, averaged over all \(B\) trees.
A large value indicates an important predictor
Similar measures/plots are calculated in boosting
Beer: Variable Importance Table
Beer: Variable Importance Plot

Boosting Comments
- Boosted models are incredibly powerful.
“best off-the-shelf classifier in the world” - Leo Breiman
- Generally speaking we have this relationship:
\[\text{Single Tree} < \text{Bagging} < \text{RF} < \text{Boosting}\]
Tuning suggestions
If learning rate \(\lambda\) is small then number of trees (\(B\)) and/or tree complexity (\(d\)) needs to increase.
Tree complexity has approximate inverse relationship with the learning rate. AKA, if you double complexity, halve the learning rate and keep the number of trees constant.
At least 1000 trees when dealing with small sample sizes (eg, \(n\) = 250) is recommended.
Summary
- Decision trees are simple and interpretable models for regression and classification
- However, they are often not competitive with other methods in terms of prediction accuracy
- Bagging, RF and boosting are good ensemble methods for improving the prediction accuracy of trees.
- The later two methods – RF and boosting – are among the state-of-the-art methods for supervised learning; however, their results can be difficult to interpret.
Comments about
gbm