Data 311: Machine Learning
Lecture 8: Bayes Classifier, KNN Classification and Discriminant Analaysis
University of British Columbia Okanagan
Introduction
Last class we saw some metrics used for assessing the models used for classification.
We have seen the logistic regression model used for binary classification (used to predict one of two possible outcomes)
-
Today we will continue with classification methods:
- Bayes Classifier
- KNN Classification
- LDA Linear Discriminant Analysis
Test Error Rate
- The test error rate associated with a set of test observations of the form (\(x_0, y_0\)) is given by
where \(\hat y_0\) is the predicted class label that results from applying the classifier to the test observation with predictor \(x_0\).
A good classifier is one for which the test error is smallest.
Bayes Classifier
“Bayes classifier” is a general concept that refers to any classifier that makes predictions based on Bayes theorem
A Bayes classifier calculates the probability of a particular class given a set of features and then selects the class with the highest probability as the prediction.
It can be shown1 that the Bayes Classifier minimizes the testing error given on the previous page.
Bayes Theorem
Bayes’ theorem relates conditional probabilities.
\[\begin{equation} P(A|B) = \frac{P(B|A) \cdot P(A)}{P(B)} \end{equation}\]\(P(A|B)\) is the conditional probability of event A occurring given that event B has occurred.
\(P(B|A)\) is the conditional probability of event B occurring given that event A has occurred.
\(P(A)\) is the marginal probability of event A
\(P(B)\) is the marginal probability of event B
Definition
The Bayes classifier assigns each observation to the most likely class, given its predictor/input values (i.e. \(X\)s ).
That is, for some predictor vector \(x_0=(x_{01}, x_{02}, \ldots, x_{0p})\) we should assign the class \(j\) where the following conditional probability is maximized: \[P(Y=j \mid X=x_0)\]
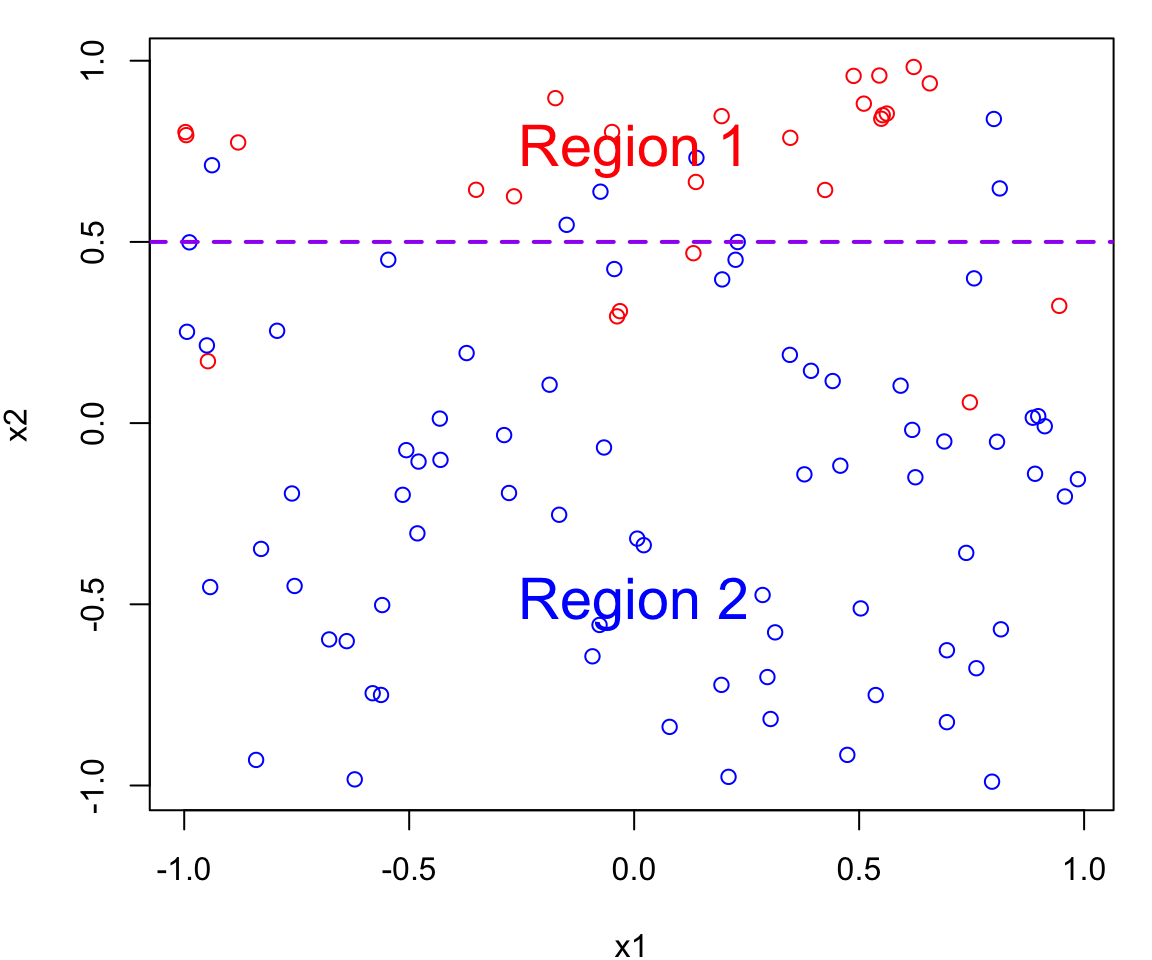
Bayes Decision Boundary Definition
For the two class problem the Bayesian decision boundary is defined as the line where
\[P(Y=1 \mid X=x_0) = P(Y=2 \mid X=x_0)\]
This boundary separates the feature space into two decision regions:
- Region 1 points for which \(P(Y = 1 \mid X = x_0) > 0.5\) and are assigned to class 1
- Region 2 points for which \(P(Y = 1 \mid X = x_0) < 0.5\) and are assigned to class 2
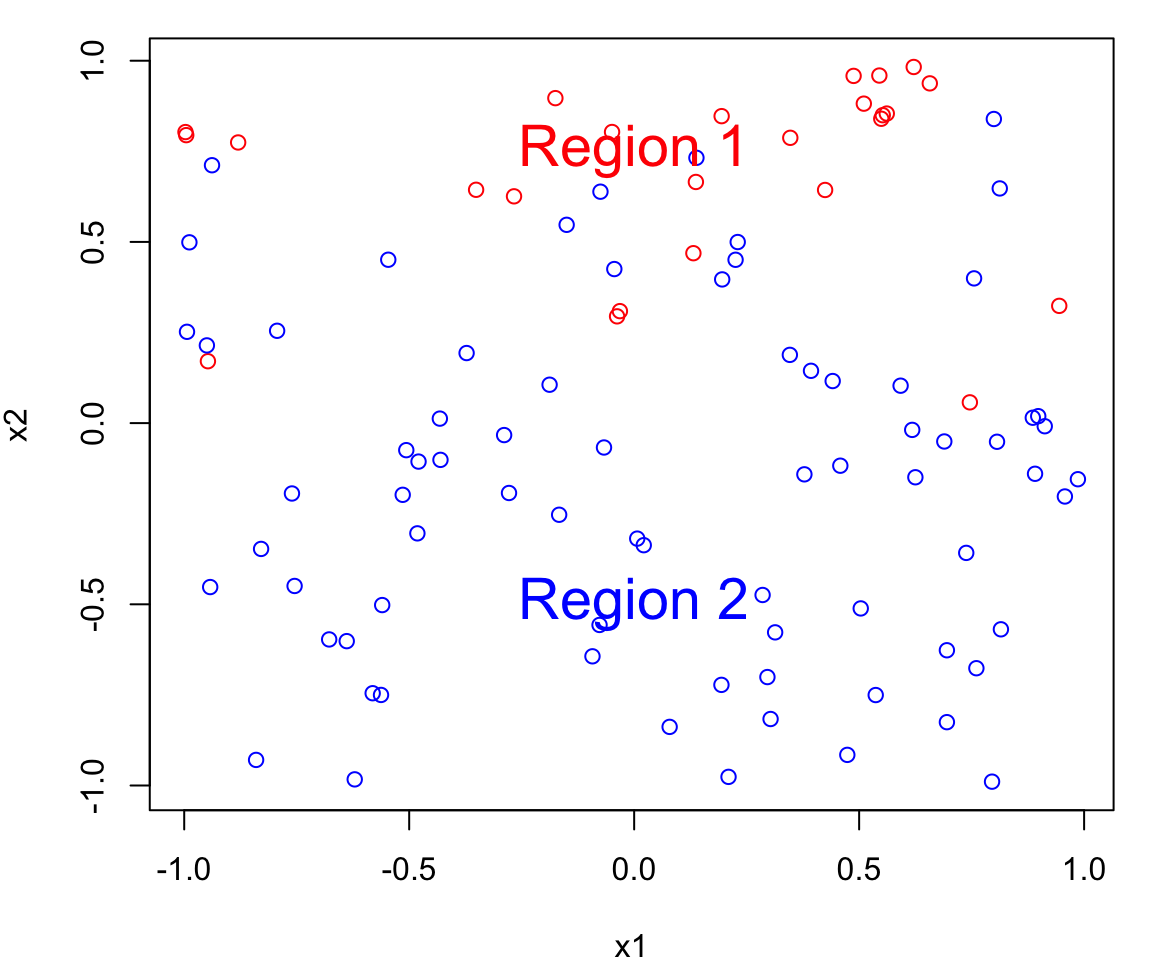
Bayes Classifier Example
Let’s simulate a data set with of two continuous predictors \(X_1\) and \(X_2\); both uniformly distributed from \([-1, 1]\).
-
We create two classes:
- a red group (corresponding to \(Y=0\)) and
- a blue group (corresponding to \(Y=1\))
To generate the class assignment we will only using \(X_2\).
That is \(X_1\) is represents a useless predictor.
Class generation
We will assign an observation to a class based on the following:
If \(x_2 > 0\)
\[\begin{align} P(Y = 0 \mid X_2 = x_2) &= x_2\\ P(Y = 1 \mid X_2 = x_2) &= 1 - x_2 \end{align}\]If \(x_2 \leq 0\)
\[\begin{align} P(Y = 0 \mid X_2 = x_2) &= 0\\ P(Y = 1 \mid X_2 = x_2) &= 1 \end{align}\]Notice that \(x_{1i}\) does not provide any information about the class.
Bayes Decision Boundary
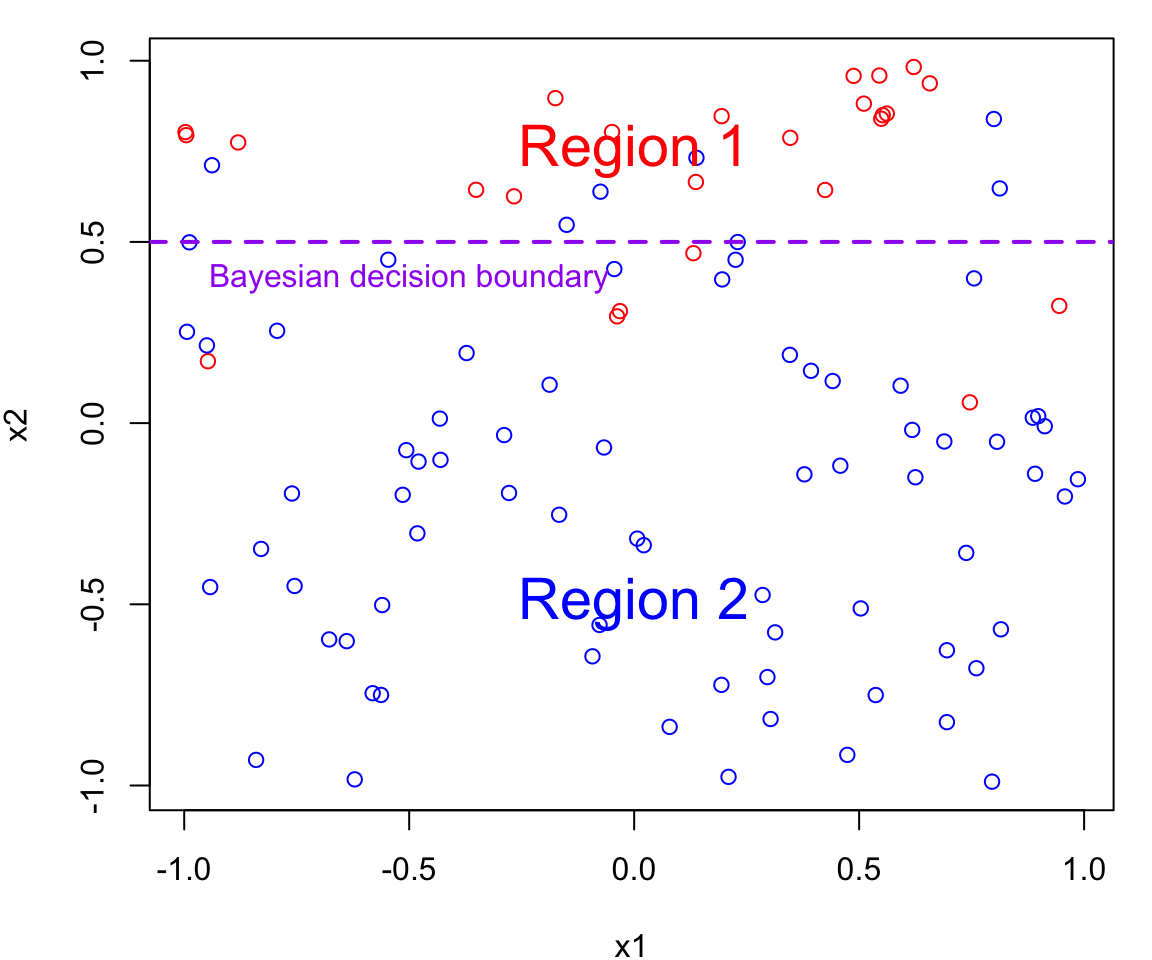
The Bayes decision boundary represents the dividing line in the feature space (i.e. in terms of \(X\)s) that determines how data points are classified into different classes.
In our case data points below the decision boundary (Region 1) are classified as red, while data points above the decision boundary (Region 2) are classified as blue
Note that errors are still going to be made using this classifier since the process is random (points near the boundary are sometimes going to red in Region 2 and sometimes blue in Region 1)
Optimal Boundary
The Bayes decision boundary represents the optimal decision boundary because it’s based on the true underlying probability distributions of the data.
In other words, the Bayes decision boundary is going to make fewer mistakes than any other classifier you will come up with.
- However, in practice, these distributions are often unknown and need to be estimated from data, which can introduce uncertainty.
Bayes Error Rate
On average, Bayes classifier will yield the lowest possible test error rate given by the following expectation (averages the probability over all possible values of \(X\)) \[1-E\left( \max_j P(Y=j \mid X) \right)\]
The above is called the Bayes error rate which represents the minimum possible error rate that any classifier can achieve for a given classification problem.
From simulations to real data
- In theory we would always like to predict qualitative responses using the Bayes classifier.
- But for real data, we do not know the conditional distribution of \(Y\) given \(X\), and so computing the Bayes classifier is impossible.
- Therefore, the Bayes classifier serves as an unattainable gold standard against which to compare other methods.
Classification Algorithms
Many approaches attempt to estimate the conditional distribution of \(Y\) given \(X\), and then classify a given observation to the class with highest estimated probability.
One example is the \(K\)-nearest neighbors (KNN) classifier.
This algorithm is similar to the KNN regression, only now, rather than averaging continuous \(y\) values, we are counting the number of neighbors that belong to each class.
The majority class among the \(K\)-nearest neighbors is chosen as the predicted class for the new data point.
KNN Classification
Given a positive integer \(K\) and a test observation \(x_0\), the KNN classifier first identifies the \(K\) points in the training data that are closest to \(x_0\), represented by \(\mathcal{N}_0\).
For each class \(j\), find \[\begin{equation} P(Y=j \mid X=x_0) \approx \frac{1}{K} \sum_{i \in N_0} I(y_i = j) \end{equation}\]
Assign observation \(x_0\) to the class (\(j\)) for which the above probability is largest
in R
To demo knn classification we will be using the knn function from the class package performs (other alternatives exist)
-
trainmatrix or data frame of training set cases. -
testmatrix or data frame of test set cases. -
clfactor of true classifications of training set -
knumber of neighbours considered.
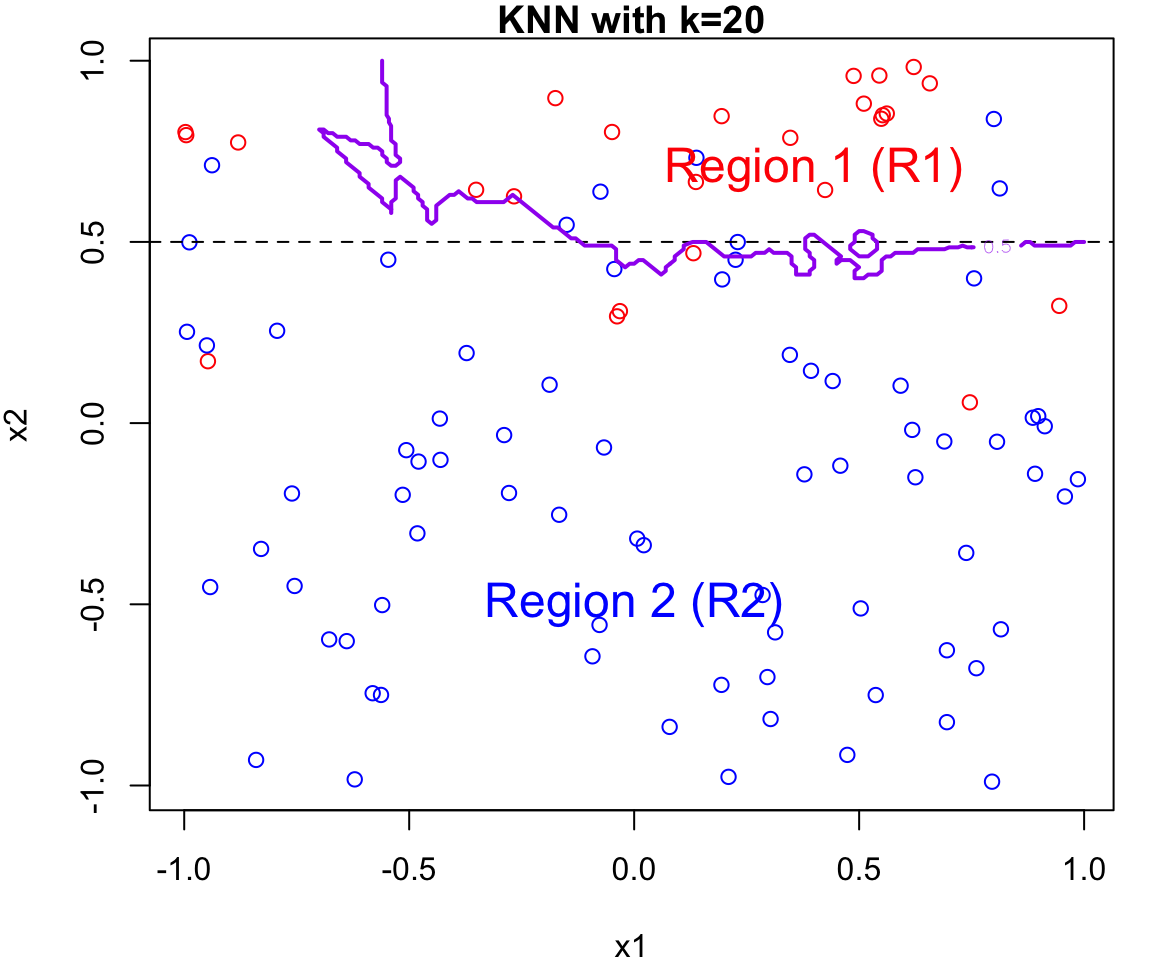
k Nearest Neighbours Example
Let’s apply this algorithm to the simulated data considered previously.
More specifically, we will fit
knnfor \(k = 20, 15, 9, 3\) and \(1\)Fair warning that the decision boundary and decision regions will not be as clear cut as our previous example.
We will assess the error rate for each (code presented in lab)




An great interactive demo can be view here
Choosing k
A key component of a “good” KNN classifier is determined by how we choose \(k\).
\(k\) in a user-defined input to our algorithm which we can set anywhere from 1 to \(n\) (the number of points in our training set)
Question What would happen if \(k\) was set equal to \(n\)?
Discuss some general rules of thumb for small/large \(k\)?
Recap
Many approaches attempt to estimate the conditional distribution of \(Y\) given \(X\), and then classify a given observation to the class with highest estimated probability.
KNN did this using a “voting” system with its nearest neighbours.
Logistic regression uses the logistic function, e.g. \(P(Y = 1 \mid X = x) = \frac{e^{\beta_0 + \beta_1x}}{1 + e^{\beta_0 + \beta_1x}}\)
Let’s discuss linear and quadratic discriminant analysis as alternative methods for approximating the conditional distribution of \(Y\) given \(X\).
Discriminant Analysis
- Here the approach is to model the distribution of \(X\) in each of the classes separately, and then use Bayes’ theorem to flip things around and obtain \(\Pr(Y |X)\).
- When we use normal (Gaussian) distributions for each class, this leads to linear or quadratic discriminant analysis.
- However, this approach is quite general, and other distributions can be used as well. We will focus on normal distributions.
Generative Models for Classification
Suppose we have qualitative response variable \(Y\) that can take on \(K\) possible distinct and unordered values.
Suppose we have two groups, that is \(Y\) can take on two possible values \(K = 1, 2\) (corresponding to group 1/2)
Let \(f_k(X) \equiv \Pr(X|Y = k)\) denote the density function of \(X\) for an observation that comes from the \(k\)th class.
Let \(\pi_k\) represent the overall or prior probability that a randomly chosen observation comes from the \(k\)th class.
Applying Bayes Theorem
Then Bayes’ theorem states that
\[\begin{equation} \Pr(Y = k|X = x) = \frac{\pi_k f_k(x)}{\sum_{l=1}^{K} \pi_l f_l(x)} \end{equation}\]This represents the posterior probability that an observation \(X = x\) belongs to the \(k\)th class.
To estimate \(p_k(x)\), and thereby approximate the Bayes classifier, we can need to estimate the \(\pi_k\)s and \(f_k(x)\).
LDA for one predictor
Let \(f(x ; \mu_k, \sigma_k)\) or \(f_k(x)\) be the probability density function (PDF) of an observation from the \(k\)th class.
To estimate \(f_k(x)\), we will first make some assumptions about its form.
The most commonly assumed PDF is the normal or Gaussian distribution with mean \(\mu\) and \(\sigma^2\) \[\begin{equation} f(x; \mu, \sigma) = \frac{1}{\sqrt{2\pi}\sigma} e^{-\frac{(x-\mu)^2}{2\sigma^2}} \end{equation}\]
Univariate Normal Distribution
Sticking with univariate for now, suppose each group is normally distributed with different means and variances
\[\begin{align*} f_1(x) &\sim N(\mu_1, \sigma_1^2)\\ f_2(x) &\sim N(\mu_2, \sigma_2^2) \end{align*}\]While discuss this method in 2-class univariate setting these concepts can be extended to the multivariate distributions with \(>2\) classes.
Plot of two univariate normals
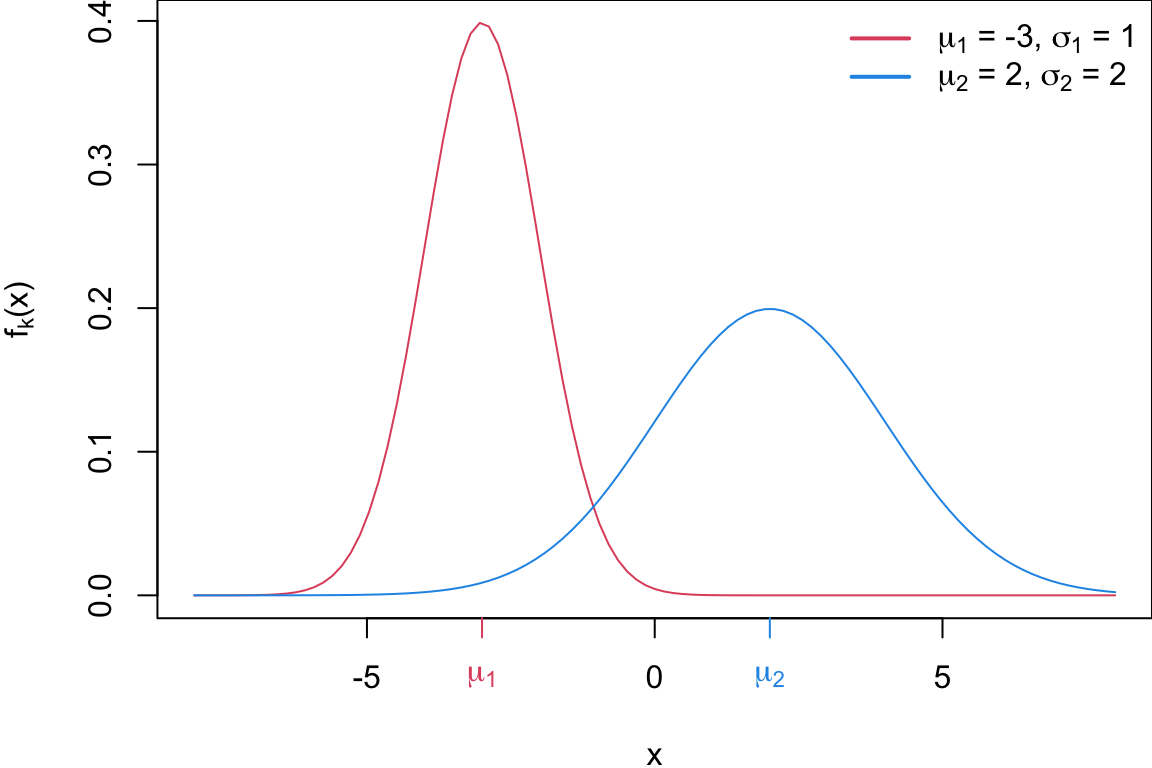
Code
par(mar = c(3.9,4.1,0,0))
pi1 = 0.6
pi2 = 1 - pi1
curve(pi1*dnorm(x, mean = mean1, sd = sd1), col = 2, from = -8, 8, ylab = expression(paste(pi[k]*f[k]*"("* x *")")), lty = 2)
curve(pi2*dnorm(x, mean = mean2 , sd = sd2), col = 4, add = TRUE, lty = 2)
axis(1, at=mean1, labels = expression(mu[1]), col=2, col.axis=2)
axis(1, at=mean2, labels = expression(mu[2]), col=4, col.axis=4)
gmm <- function(x, pis, mus, sigs){
pis[1]*dnorm(x, mean = mus[1], sd = sigs[1]) +
pis[2]*dnorm(x, mean = mus[2], sd = sigs[2])
}
curve(gmm(x, c(pi1, pi2), c(mean1, mean2), c(sd1, sd2)), col = 3, add = TRUE)
ltext <- c(bquote(mu[1] *" = "* .(mean1)*", "*sigma[1] *" = "* .(sd1)),
bquote(mu[2] *" = "* .(mean2)*", "*sigma[2] *" = "* .(sd2)),
bquote(pi[1] *" = "* .(pi1)*", "*pi[2] *" = "* .(pi2)))
legend("topright", lwd= 2, col = c(2,4, 3),
legend = ltext, bty = "n")
Building a classifier
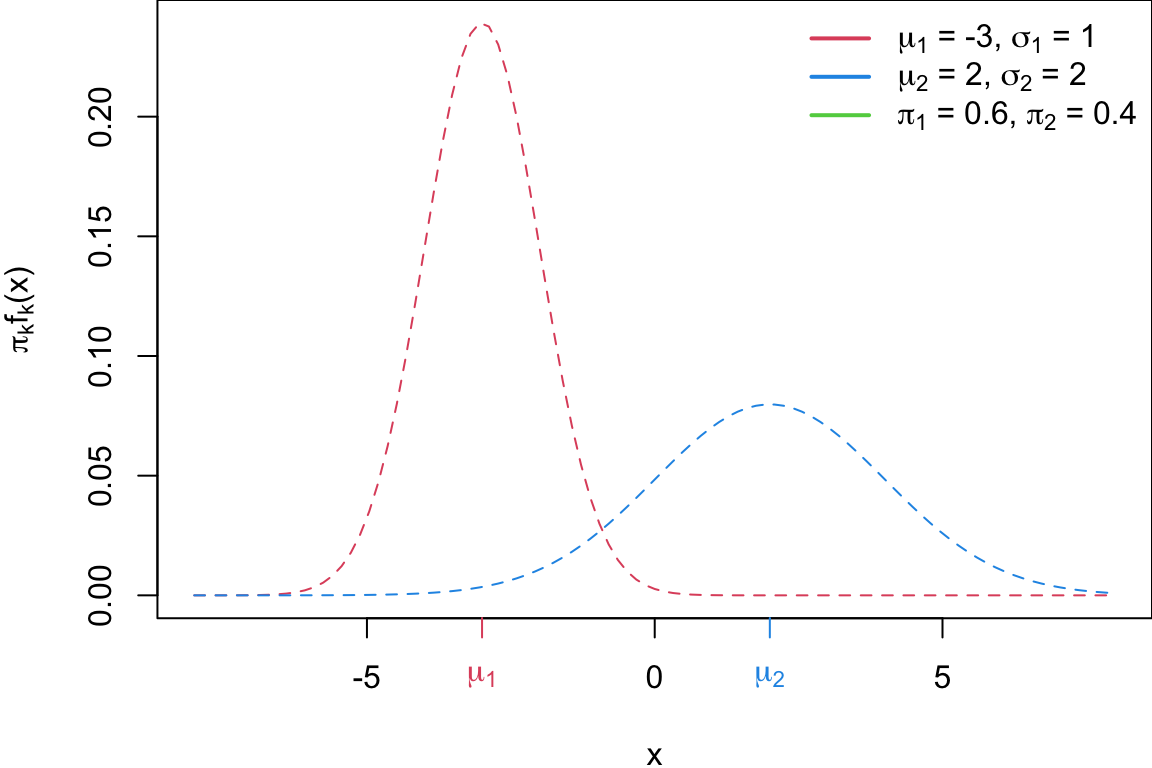
We classify new points according to whichever numerator \((\pi_k f_k(x))\) is the highest.
Question: where would that decision boundary be on this picture?
LDA
Linear Discriminant Analysis, or LDA for short, assumes that \(\sigma_1 = \sigma_2\)
When we relax this assumption and allow for differing variances between groups, we get Quadratic Discriminant Analysis, or QDA for short
For simplicity, we will focus on the LDA univariate case first with equal priors \(\pi_1 = \pi_2\)
We will then focus on univariate LDA with equal priors \(\pi_1 = \pi_2\) and extend the ideas to multivariate QDA.

If the class variances and priors are equal, the decision boundary is simply the average of the means

If the class variances and different and the priors are equal, the decision boundary is a function of sigma, the priors, and the means
LDA decision boundary
Since we’re dealing with two classes, we can set the decision boundary where \(p_1(x) = p_2(x)\)
\[\begin{align} \frac{\pi_1 f_1(x)}{\sum_{l=1}^{K} \pi_l f_l(x)} &= \frac{\pi_2 f_2(x)}{\sum_{l=1}^{K} \pi_l f_l(x)} \\ \frac{\pi_1}{\sqrt{2\pi}\sigma} e^{-\frac{(x-\mu_1)^2}{2\sigma^2}} &= \frac{\pi_2}{\sqrt{2\pi}\sigma} e^{-\frac{(x-\mu_2)^2}{2\sigma^2}}\\ \vdots \end{align}\]Discriminant score
This is boils down to to assigning the observation to the class for which discriminant score \(\delta_k(x)\) is the largest: \[\begin{equation} \delta_k(x) = x \cdot \frac{\mu_k}{\sigma^2} - \frac{\mu_{k}^2}{\sigma^2} + \log(\pi_k) \end{equation}\]
Decision boundary
The Bayes decision boundary when \(\pi_1 = \pi_2\) is found at: \[\begin{equation} x = \frac{\mu_1^2 - \mu_2^2}{2(\mu_1 - \mu_2)} = \frac{\mu_1 + \mu_2}{2} \end{equation}\]
Let’s find the Bayes decision boundary when \(\pi_1 \neq \pi_2\) …
Probability of each class
Where \(\pi_1\) = 0.8, \(\pi_2\) = 0.2, \(\sigma = \sigma_1 = \sigma_2\) = 2, \(\mu_1\) = -3 \(\mu_2\) = 2 and x = 0.61
\[ \begin{align} P(&Y = 1 \mid x = 0.61) \\ &= \frac{\pi_1 f_1(x)}{\pi_1 f_1(x) + \pi_2 f_2(x)}\\ &= \frac{0.8 \cdot 0.04} {0.8 \cdot 0.04 + 0.2 \cdot 0.16}\\ &= 0.5 \end{align} \]
Why discriminant analysis
- If \(n\) is small and the distribution of the predictors \(X\) is approximately normal in each of the classes, the linear discriminant model is more stable than the logistic regression model.
- When the classes are well-separated, the parameter estimates for the logistic regression model are surprisingly unstable. Linear discriminant analysis does not suffer from this problem.
- LDA is popular when we have more than two response classes, because it also provides low-dimensional views of the data (we’ll return to this idea later)
Classifier
When we do not have equal variance (i.e \(sigma_1 \neq \sigma_2\)) our classifier predicts \(x_0\) to group 1 if
\[ \pi_1 \cdot f_1(x; \mu_1, \sigma_1) > \pi_2\cdot f_2(x; \mu_2, \sigma_2) \]
More generally, we classify observation \(x\) based on the following rule:
\[ \hat y = \underset{k}{\operatorname{argmax}} \pi_k \cdot f_k(x; \mu_k, \sigma_k) \]
But we don’t know \(f_k\)
No problem. We estimate them!

ILSR2 Figure 4.4: Left: Two one-dimensional normal density functions are shown. The dashed vertical line represents the Bayes decision boundary. Right: 20 observations were drawn from each of the two classes, and are shown as histograms. The Bayes decision boundary is again shown as a dashed vertical line. The solid vertical line represents the LDA decision boundary estimated from the training data.
Parameter Estimates
\[\begin{align} \hat{\pi}_k &= \frac{n_k}{n} \quad\quad \hat{\mu}_k = \frac{1}{n_k} \sum_{i: y_i=k} x_i\\ \hat{\sigma}^2 &= \frac{1}{n - K} \sum_{k=1}^{K} \sum_{i: y_i=k} (x_i - \hat{\mu}_k)^2 \end{align}\]where \(n\) is the total number of training observations, \(n_k\) is the number of observations in class \(k\), and \(\mu_k\) and \(\hat{\sigma}^2_k\) are the MLE estimates for mean and (pooled) variance in the \(k\)th class.
Multivariate Normal Distribution
For Discriminant analysis when \(p>1\) we use the Multivariate Normal Distribution (MVN).
A MVN random vector \(\mathbf X\), has probability distribution given by \[f(\mathbf x) = \frac{1}{(2\pi)^{p/2}|\boldsymbol{\Sigma}|^{1/2}} e^{-\frac{1}{2}(\mathbf{x}-\boldsymbol{\mu})^T\boldsymbol{\Sigma}^{-1}(\mathbf{x}-\boldsymbol{\mu})}\] with mean vector \(\boldsymbol{\mu}\) and covariance matrix \(\boldsymbol{\Sigma}\).
3D Plot of Multivariate Normal

Covariance Matrix
The covariance matrix is symmetric a square matrix where each entry \(\Sigma_{ij}\) represents the covariance between variables \(X_i\) and \(X_j\)
The diagonal elements (\(\sigma_{i}\)) represent the variances of individual variables, and the off-diagonal elements (\(\sigma_{ij}\) for \(i\neq j\)) represent the covariances between pairs of variables. \[\begin{equation} \boldsymbol \Sigma = \begin{bmatrix} \sigma_{1}^2 & \sigma_{12} & \ldots & \sigma_{1p} \\ \sigma_{12} & \sigma_{2}^2 & \ldots & \sigma_{2p} \\ \vdots & \vdots & \ddots & \vdots \\ \sigma_{1p} & \sigma_{2p} & \ldots & \sigma_{p}^2 \end{bmatrix} \quad \text{ where }\sigma_{ij} = \sigma_{ji} = \text{Cov}(X_i, X_j) \end{equation}\]
Uncorrelated
Code
# Install and load the required libraries
library(plotly)
library(mvtnorm)
# Parameters for the multivariate normal distribution
mean_vec <- c(0, 0) # Mean vector
cov_mat <- matrix(c(1, 0, 0, 1), nrow = 2) # Covariance matrix
# Generate data grid
x <- seq(-3, 3, length.out = 100)
y <- seq(-3, 3, length.out = 100)
grid <- expand.grid(x, y)
# Calculate the multivariate normal density for each point in the grid
z <- dmvnorm(grid, mean = mean_vec, sigma = cov_mat)
# Reshape the z values to match the grid dimensions
z_matrix <- matrix(z, nrow = length(x), ncol = length(y))
# Create an interactive 3D surface plot using plotly
plot_ly(z = ~z_matrix, x = ~x, y = ~y, type = "surface") %>%
layout(scene = list(
xaxis = list(title = "X-axis"),
yaxis = list(title = "Y-axis"),
zaxis = list(title = "Density")
))Code
# Install and load the required libraries
library(plotly)
library(mvtnorm)
# Parameters for the multivariate normal distribution
mean_vec <- c(0, 0) # Mean vector
cov_mat <- matrix(c(1, 0.5, 0.5, 1), nrow = 2) # Covariance matrix
# Generate data grid
x <- seq(-3, 3, length.out = 100)
y <- seq(-3, 3, length.out = 100)
grid <- expand.grid(x, y)
# Calculate the multivariate normal density for each point in the grid
z <- dmvnorm(grid, mean = mean_vec, sigma = cov_mat)
# Reshape the z values to match the grid dimensions
z_matrix <- matrix(z, nrow = length(x), ncol = length(y))
# Create an interactive 3D surface plot using plotly
plot_ly(z = ~z_matrix, x = ~x, y = ~y, type = "surface") %>%
layout(scene = list(
xaxis = list(title = "X-axis"),
yaxis = list(title = "Y-axis"),
zaxis = list(title = "Density")
))LDA when \(p>1\)
In the case of \(p > 1\) predictors, the LDA classifier assumes that the observations in the \(k\)th class are drawn from a multivariate Gaussian distribution \(\mathcal{N}(\boldsymbol\mu_k, \boldsymbol\Sigma)\), where \(\boldsymbol\mu_k\) is a class-specific mean vector, and \(\boldsymbol\Sigma\) is a covariance matrix that is common to all \(K\) classes.
Algebra reveals that the Bayes classifier assigns an observation \(X = x\) to the class for which \[\begin{equation} \delta_k(x) = x^T \boldsymbol\Sigma^{-1} \boldsymbol\mu_k - \frac{1}{2} \boldsymbol\mu_k^T \boldsymbol\Sigma^{-1} \boldsymbol\mu_k + \log(\pi_k) \end{equation}\]
What does equal Covariance look like?

Source of image [1]
Contour Plots (equal covariance)
Code
# Load the required libraries
library(ggplot2)
library(mvtnorm)
library(cowplot)
# Set a random seed for reproducibility
set.seed(123)
# Define the means and covariance matrix
mean1 <- c(2, 3) # Mean vector for distribution 1
mean2 <- c(4, 4) # Mean vector for distribution 2
cov_matrix <- matrix(c(1, 0.5, 0.5, 1), nrow = 2) # Shared covariance matrix
# Generate data grid
x <- seq(0, 6, length.out = 400)
y <- seq(0.5, 6.5, length.out = 400)
grid <- expand.grid(x, y)
# Calculate the density for each point in the grid for both distributions
z1 <- dmvnorm(grid, mean = mean1, sigma = cov_matrix)
z2 <- dmvnorm(grid, mean = mean2, sigma = cov_matrix)
# Combine the densities into a data frame
data1 <- data.frame(x = rep(x, length(y)), y = rep(y, each = length(x)), z = z1)
data2 <- data.frame(x = rep(x, length(y)), y = rep(y, each = length(x)), z = z2)
ggplot() +
geom_contour(data = data1, aes(x = x, y = y, z = z), color = "blue", bins = 8) +
geom_contour(data = data2, aes(x = x, y = y, z = z), color = "red", bins = 8) +
labs(title = "Contours for Two Functions") +
theme_minimal()
Illustration \(p = 2\) and \(K=3\)

ILSR Fig 4.6: Here \(\pi_1 = \pi_2 = \pi_3\) Left: Ellipses that contain 95 % of the probability for each of the three classes are shown. The dashed lines are the Bayes decision boundaries. Right: 20 observations were generated from each class, and the corresponding LDA decision boundaries are indicated using solid black lines. The Bayes decision boundaries are shown as dashed lines.
LDA to QDA
When \(f_k(x)\) are Gaussian densities, with the same covariance matrix \(\boldsymbol\Sigma\) in each class, this leads to linear discriminant analysis.
With Gaussians but different \(\boldsymbol \Sigma_k\) in each class, we get quadratic discriminant analysis.
Because the \(\boldsymbol\Sigma_k\) are different, we don’t loose the quadratic terms so we get discriminant function is now
Contour Plots (unequal covariance)
Code
# Load the required libraries
library(ggplot2)
library(mvtnorm)
library(cowplot)
# Set a random seed for reproducibility
set.seed(123)
# Define the means and covariance matrix
mean1 <- c(2.5, 3)
mean2 <- c(3, 3)
# Define different covariance matrices for the two distributions
cov_matrix1 <- matrix(c(1, 0.3, 0.3, 2), nrow = 2)
cov_matrix2 <- matrix(c(2, -0.4, -0.4, 1), nrow = 2)
# Generate data grid
x <- seq(-1, 6, length.out = 400)
y <- seq(-1, 6.5, length.out = 400)
grid <- expand.grid(x, y)
# Calculate the density for each point in the grid for both distributions
z1 <- dmvnorm(grid, mean = mean1, sigma = cov_matrix1)
z2 <- dmvnorm(grid, mean = mean2, sigma = cov_matrix2)
# Combine the densities into a data frame
data1 <- data.frame(x = rep(x, length(y)), y = rep(y, each = length(x)), z = z1)
data2 <- data.frame(x = rep(x, length(y)), y = rep(y, each = length(x)), z = z2)
ggplot() +
geom_contour(data = data1, aes(x = x, y = y, z = z), color = "blue", bins = 5) +
geom_contour(data = data2, aes(x = x, y = y, z = z), color = "red", bins = 5) +
labs(title = "Contours for Two Functions") +
theme_minimal()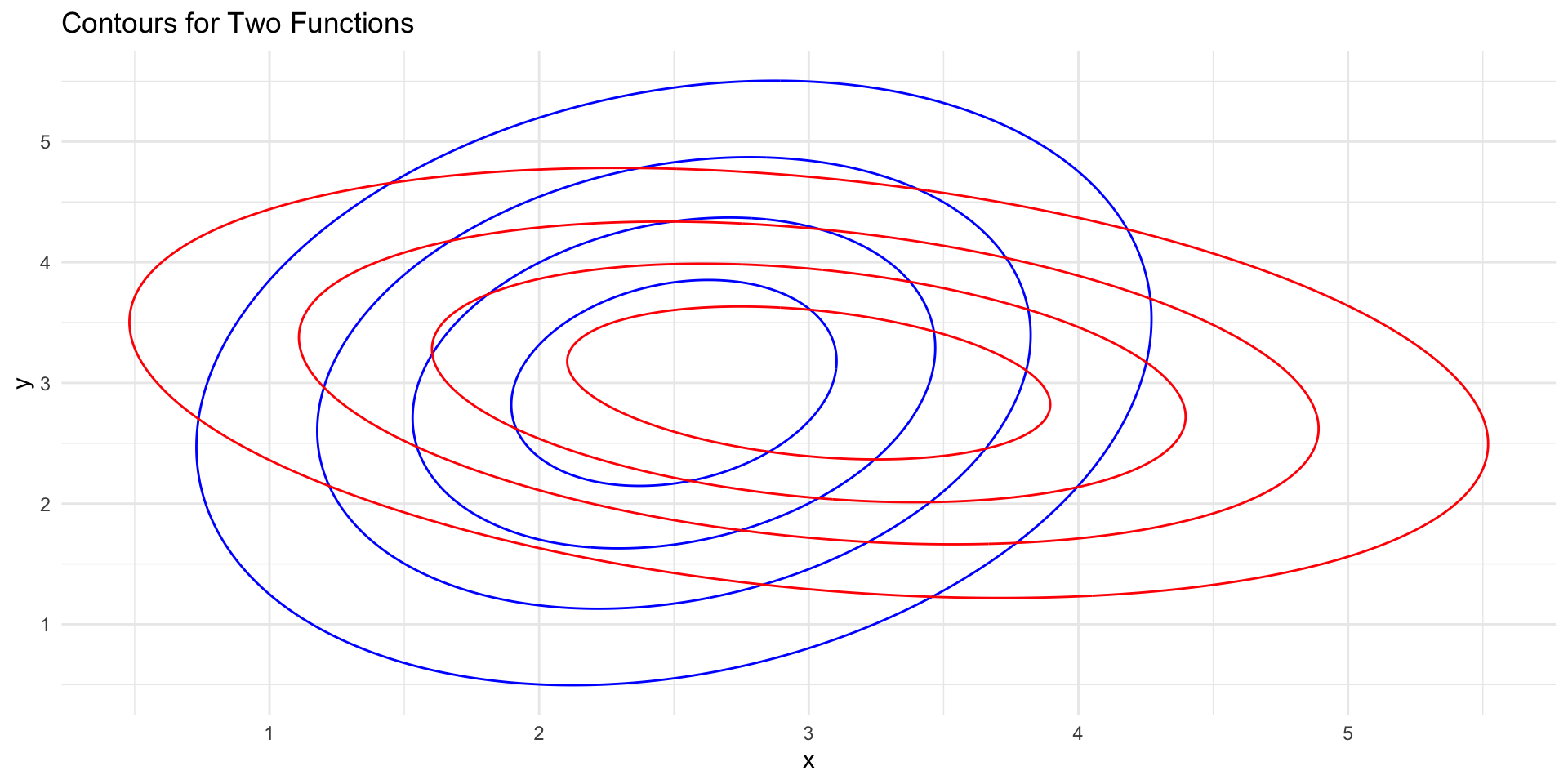
Summary
Both LDA and QDA assumes each class follows an underlying Gaussian distribution
LDA
assumes equal variance across classes
PRO: easier to fit
CON: less flexible
QDA
doesn’t assume equal variance across classes
PRO: often leads to a lower error rate
CON: reduced intelligibility
Bias-variance tradeoff: the LDA can be too simple whereas the QDA has a higher risk of overfitting to our data.
Example: Iris
LDA fitted model
Let’s fit the LDA classifier using all 4 predictors
...
Prior probabilities of groups:
setosa versicolor virginica
0.3333333 0.3333333 0.3333333
Group means:
Sepal.Length Sepal.Width Petal.Length Petal.Width
setosa 5.006 3.428 1.462 0.246
versicolor 5.936 2.770 4.260 1.326
virginica 6.588 2.974 5.552 2.026
...Plotted fit

This of course is the training error so it may be overly optimistic.
Classification Table
LDA classifies all but 3 of the 150 training samples correctly.
From \(\delta_k(x)\) to probabilities
From \(\delta_k(x)\) to probabilities, once we have estimates \(\hat{\delta}_k(x)\), we can turn these into estimates for class probabilities:
\[\begin{align} \Pr(Y = k \,|\, X = x) = \frac{e^{\hat{\delta}_k(x)}}{\sum_{l=1}^{K} e^{\hat{\delta}_l(x)}} \end{align}\]So classifying to the largest \(\hat{\delta}_k(x)\) amounts to classifying to the class for which \(\Pr(Y = k \,|\, X = x)\) is largest.
