3 + 4[1] 73 - 4[1] -13/4[1] 0.753^2[1] 9pi^2[1] 9.869604An Introduction to R and R markdown
Dr. Irene Vrbik
September 18, 2023
Summary This lab will outline some of the basics in R, as well as introduce R markdown as a file format for making dynamic documents for assignments. This lab assignment is not for marks. In the future, your assignments will involve creating Rmd files and submitting the resulting HTML outputs as your work. However, for this lab, there’s no need to submit anything. If you are confident in your ability to achieve the learning outcomes, proceed directly to the exercises and confirm that you can complete them successfully.
By the end of this lab students will be able to:
In DATA 311, we will use R (and RStudio) in lab and for completing assignments. R is free, open-source, and multi-platform, so you should be able to install it on your machine of choice. RStudio is proprietary, but has free-for-personal-use versions, and can be used on all major platforms — Windows, Mac, Linux.
If you are unable to get Rstudio, you are free to use an alternative method for creating readable assignments (eg. word documents, Jupyter notebook) but it is highly recommended that you use Rstudio. If you are using a chromebook for example, there are workarounds for getting Rstudio installed (eg first two google searches: 1, 2)
To complete all the steps of this lab you will need to do the following:
Even if you have these programs installed on your computer you should update them to ensure that you have the latest version.
Much of R works as you might expect. It can function as a basic calculator…
You can make objects and assign values…
Note that in R you can use <- or = as the assignment operator. Namely, we could have written:
R is case-sensitive
And be careful about leaving out operators…
versus
We can create a vector using c (for combine)
Vectors can contain strings instead of numbers:
We can also make quick sequences…
[1] 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22
[19] 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40
[37] 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58
[55] 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76
[73] 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94
[91] 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112
[109] 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130
[127] 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148
[145] 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166
[163] 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184
[181] 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202
[199] 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220
[217] 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238
[235] 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256
[253] 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274
[271] 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292
[289] 293 294 295 296 297 298 299 300We can check how many elements a vector has using lenght() function:
To index an element from a vector, use single square brackets
To index multiple elements from a vector we could use:
[1] "bananas" "oranges"[1] "pineapples" "bananas" [1] "bananas" "oranges" "pineapples"We can also extract elements of our vector that fulfill a certain condition. For example let’s extract all of the elements that are larger than 3 from vec
Note that vec>3 is a logical vector:
This logical vector in this case is being used to index all the elements for which this logical check returns true. In this case, we are extracting elements 2, 3, 4. To extract that index in R we could use the which() function
Checks can also be done on strings. For example,
[1] "apples"[1] "apples" "pineapples"There is almost always more than one way of accomplishing a task. For example, finding the average of our vector vec
[1] 3.5[1] 3.5[1] 3.5[1] 3.5Notice the use of # to indicate the start of a comment.
For another example, how about the standard deviation of vec?
[1] 1.870829[1] 1.870829But without having manually calculated the sample standard deviation beforehand, how could we be confident that the “sd” function is using the unbiased divisor “(n - 1)”? Type the following in your console (bottom left window under default RStudio settings), then hit enter:
You should see a help document pop up in the bottom right window (again, default Rstudio settings). If you read through the Details section of that document, it will specify the denominator it is using.
Caution: R is open-source, and even packages hosted on the official CRAN repository will vary in terms of the actual helpfulness of the help files.
By default, matrices are constructed columnwise.
You can always change to row-wise by specifying the argument byrow=TRUE
The following extracts a column, row, and cell from matrix m,
[1] 3 6[1] 1 2 3[1] 3Data frames are perhaps the most used data structure used in statistics. They can store different data types and can contain additional attributes such as column and row names. When combining vectors in a data frame all must have the same size (i.e. length). Missing observations will be recorded as NA.
Here is an example of how to create a data frame and add to it:
Person Grade
1 John 45
2 Jill 92
3 Jack 91To extract the first column, we can either reference it by number:
OR we can reference it by name:
A third option is to extract a column of this data frame using the $ operator:
Adding a column to the data frame is as easy as typing:
Alternatively, we could have let R figure out whether the student passed or not based on the grade and typed:
Many data sets are available in “base” R and a larger collection can be accessed via R packages. For example, if we want to gain access to the Old Faithful data frame, we could type:
To learn more on this data set type ?faithful into the R console. To see the first few rows of this data set type:
Another cool feature is we can get a summary of each column using:
eruptions waiting
Min. :1.600 Min. :43.0
1st Qu.:2.163 1st Qu.:58.0
Median :4.000 Median :76.0
Mean :3.488 Mean :70.9
3rd Qu.:4.454 3rd Qu.:82.0
Max. :5.100 Max. :96.0 As before, we can extract column names using the dollar sign and perform operations on those vectors, for example
Rather than calling the columns from the data frame using $, you could instead “attach” the data set (see ?attach). This essentially means the columns will be saved to variables that you can call directly. For example, the following code will produce an error since,
However, once we attach the faithful data set, R will have a vector called eruptions and waiting:
In a practical setting, we will most likely be required to load our own data into R at some point. The easiest way to do this is by saving your data in csv (comma separated values) format and using the read.csv() function; see ?read.csv for more details. Assuming your data is stored in your working directory (more on this in a second), you can load your csv file into R using the read.csv function:
read.csv(name_of_file.csv).
Typically, we would like to save the data set to some object that can perform actions on. For example, I can load a simple data matrix (stored in file datamatrix.csv) and save it to an object called dat:
gender sleep intro_extra countries dread
1 male 5.0 extravert 3 3
2 female 7.0 extravert 7 2
3 female 5.5 introvert 1 4
4 male 7.0 extravert 2 2
5 other 3.0 introvert 1 3
6 female 3.0 extravert 9 4The above code assumes you data is stored in you working directory. Read on to see how to specify this in R. This data set is available on Canvas for you to test.
Alternatively, we can pull a csv file straight from the web:
X species island bill_length_mm bill_depth_mm flipper_length_mm
1 1 Adelie Torgersen 39.1 18.7 181
2 2 Adelie Torgersen 39.5 17.4 186
3 3 Adelie Torgersen 40.3 18.0 195
4 4 Adelie Torgersen NA NA NA
5 5 Adelie Torgersen 36.7 19.3 193
6 6 Adelie Torgersen 39.3 20.6 190
body_mass_g sex year
1 3750 male 2007
2 3800 female 2007
3 3250 female 2007
4 NA <NA> 2007
5 3450 female 2007
6 3650 male 2007Independent of R, it is usually a good idea to put school projects into some organized folder system. For instance, in my Documents folder on my Mac I currently have the following filing organization:
data311/
|-- labs/
| |-- Lab00.Rmd
| |-- practice00.Rmd
| |-- datamatrix.csv
|-- assignemnts/
| |-- template.Rmd
| |-- example.csvSuppose I am working on this lab and I want to read in the datamatrix.csv file. I could access the file using the whole path. For example:
but I don’t recommend this (and if fact this won’t work within Rmd documents–more on this below).
Alternatively (and preferably) you should save this into your working directory. The working directory is just a file path on your computer that sets the default location of any files you read (or write out) into R. To set your working directory use setwd. Following from the file organization example above, if I want my working directory to be the labs folder, I would need to specify that path as the sole argument in the setwd function:
Once you set your working directory to the same folder in which your data file is stored you can reference it with no path.
N.B. You can check what your working directory is use getwd(). If you are working within an R Markdown document your working directory is the folder that contains the Rmd file (more on this below)
Most standard statistical analyses are built-in, but a vast array of more complex analyses are available. We will often require installation of additional packages to complete assignments and labs. For example, if you want to install the ISL2 package (the package associated with our text book) type:
The above needs only to be done once. With every new R session/script/Rmd file, if you want to gain access to all of the contents within this package, we first need to load or “attach” it using:
By loading this library into our session, we can now access any functions and data sets within this package. The manual and details can be found on the CRAN website: https://cran.rstudio.com/web/packages/ISLR2/index.html
Let’s learn the structure of writing our own functions in R. To learn the syntax, we’ll simply make a function that adds 10 to any inputted value.
Note that the final line in the function will be the outputted value. Let’s test it:
In many of the ‘artistic’ olympic sports, the judging panel is comprised of several countries, and any participant’s score is averaged (or totaled) after removing both the minimum and maximum values. Let’s write a function in R which performs this type of averaging for any inputted vector of numeric values. Call it olymean.
Let’s test out this function on the ficticious data scores in the cheer.csv file which contains the scores for two Cheerleeding teams (Navarro and Trinity Valley) across five judges. First let’s read in the data set and view it:
Judge Navarro Trinity.Valley
1 1 9.8 9.9
2 2 9.6 9.9
3 3 9.7 9.8
4 4 9.7 9.7
5 5 9.9 9.5N.B. spaces are automatically replaced by . in column names since R does not allow spaces in variable names.
[1] 9.74[1] 9.733333[1] 9.74[1] 9.8What happens if your input data includes missing values? While these are not scores, let’s try and apply this function to the Ozone values in the airquality data set which indeed contain NAs (i.e. missing values)
[1] 41 36 12 18 NA 28 23 19 8 NA 7 16 11 14 18 14 34 6
[19] 30 11 1 11 4 32 NA NA NA 23 45 115 37 NA NA NA NA NA
[37] NA 29 NA 71 39 NA NA 23 NA NA 21 37 20 12 13 NA NA NA
[55] NA NA NA NA NA NA NA 135 49 32 NA 64 40 77 97 97 85 NA
[73] 10 27 NA 7 48 35 61 79 63 16 NA NA 80 108 20 52 82 50
[91] 64 59 39 9 16 78 35 66 122 89 110 NA NA 44 28 65 NA 22
[109] 59 23 31 44 21 9 NA 45 168 73 NA 76 118 84 85 96 78 73
[127] 91 47 32 20 23 21 24 44 21 28 9 13 46 18 13 24 16 13
[145] 23 36 7 14 30 NA 14 18 20By default, we will get an error when we try to calculate the average of these values:
If we look at the documentation for the mean function (?mean) you will see that there is an argument na.rm that tells R whether NA vluaes should be removed before the computation proceeds. By default this argument set to FALSE. If, we change this to TRUE, R will calculate the average with the NAs removed:
Interestingly, our olymean function works fine:
This is because the sort function removed NAs by default (see ?sort):
[1] 1 4 6 7 7 7 8 9 9 9 10 11 11 11 12 12 13 13
[19] 13 13 14 14 14 14 16 16 16 16 18 18 18 18 19 20 20 20
[37] 20 21 21 21 21 22 23 23 23 23 23 23 24 24 27 28 28 28
[55] 29 30 30 31 32 32 32 34 35 35 36 36 37 37 39 39 40 41
[73] 44 44 44 45 45 46 47 48 49 50 52 59 59 61 63 64 64 65
[91] 66 71 73 73 76 77 78 78 79 80 82 84 85 85 89 91 96 97
[109] 97 108 110 115 118 122 135 168Since we are calling sort before calling the mean function this will run without error, however, it is important to note that this function is removing the smallest number, the largest number, AND any missing values before computing the mean.
R Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents; see http://rmarkdown.rstudio.com. This lab1 is an example of an R Markdown document created using the RMarkdown R package. You can install the necessary packages using:
This section of the lab will help you from going from an .Rmd as shown on the left-hand side of figure below to a html or pdf document (which is the document you are currently viewing!).
R markdown files support basic markdown (a “lightweight” markup language) syntax with the additional benefit of allowing you to include R code and output (generated from the embedded R code) directly within the same document. Please read through this markdown cheatsheet to get familiar with the simple syntax. For example, we use _italics_ for italics, **bold** for bold, # to denote level 1 headers, ## for level 2 headers, etc… We can create simple lists
Numbered lists
and Tables:
| Tables | Are | Cool |
|---|---|---|
| left-aligned | centered | right-aligned |
| more | data | here |
.Rmd fileTo create an R markdown file, click New File > R Markdown. In the pop-up window you can specify a name, author, and output format.
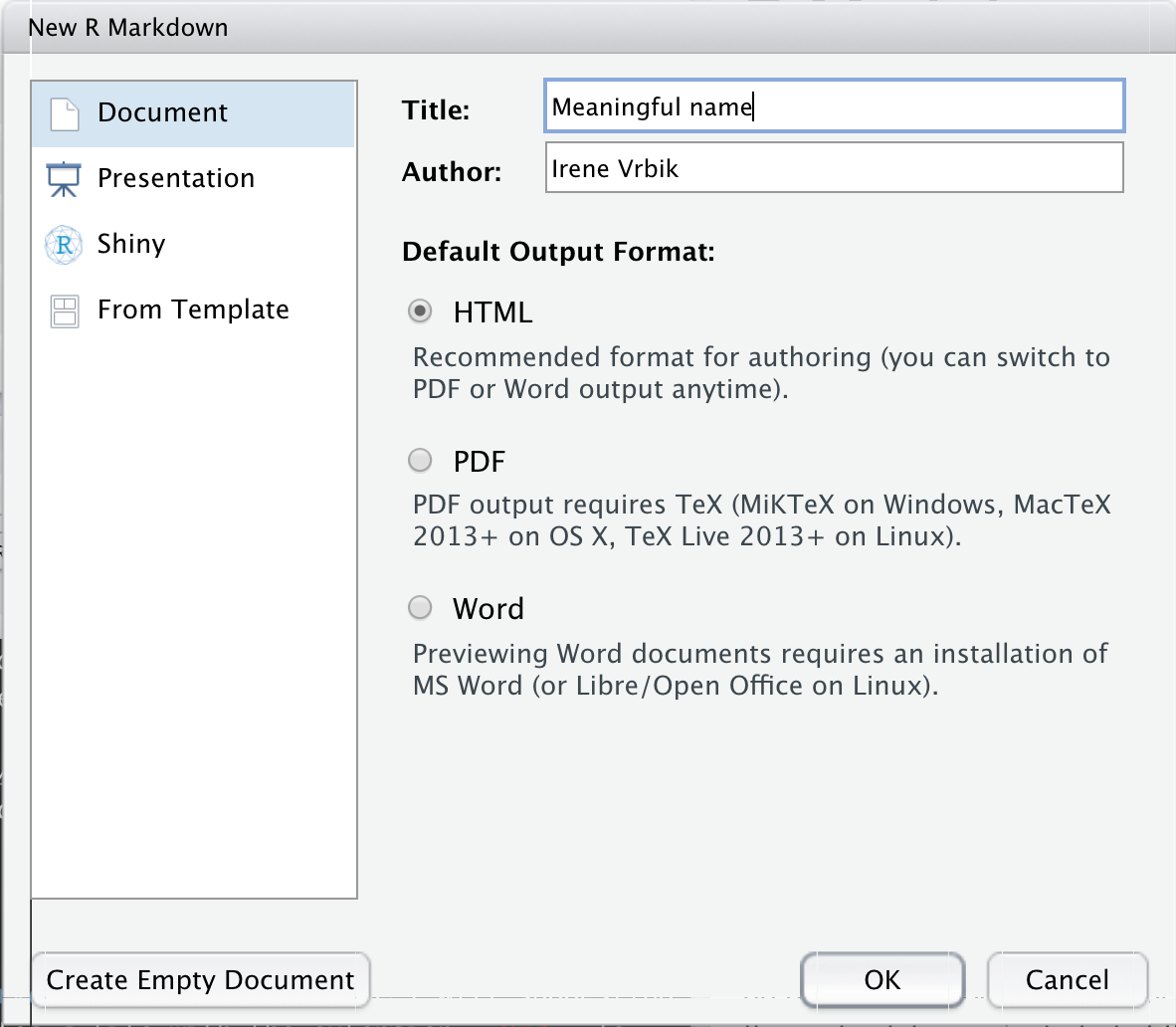
For this course I will suggesting that you create HTML documents. Press the Create Empty Document button if you would do not want your file to have any auto-populated text (although when you are first starting out you may find the example text helpful).
Warning In order to knit to pdf documents you must have installed a LaTex distribution in your system. Similarly, in order to knit to a Word Document, you will need Microsoft Word (or a similar program) installed. You can read about install LaTex in Ch 1 of the R Markdown: The Definitive Guide book. Knitting to PDF/Word will not be necessary for this course so please feel free to stick to knitting to HTML for simplicity.
One you have pressed “OK” it will open an Untitled R markdown file. At the very top of the document you will see the YAML (which stands for Yet Another Markup Language). It contains the meta-data used to configure of your document. In it’s simplest form you should have the following information in your YAML
---
title: "something meaninful"
author: "FULL name and student number"
date: "date created or last edited are reasonable"
output: html_document
---To complete your assignments you can edit this file, or download and use the assignment template that I have uploaded to Canvas (template.Rmd). Please use a more meaningful name than Untitled.Rmd or template.Rmd. Notably, I will usually be asking you to submit the rendered html document to Canvas for grading. That is, the source .Rmd file will not be required for submission unless otherwise requested.
TIP Knit as you you go! If you leave the knitting to the very end, you will often get cryptic error messages and it will be impossible to pin point where you went wrong. I tend to knit every time a paragraph or new chunk is created. That way, if an error happens, I know exactly where in my .Rmd file I should concentrate on debugging.
.Rmd fileThe creator of knitr Yihui Xie designed the package to be an “engine for dynamic report generation with R”. In order to convert your .Rmd file into a more readable html or pdf document, we will need to render AKA compile AKA knit it in RStudio. The easiest way to do this is to click the Knit button (located in the Editor window–look for the yarn and needle icon) or to use the keyboard shortcut Ctrl + Shift + K (Cmd + Shift + K on macOS). When you “knit”, either a html or pdf document will be generated that converts any markdown syntax into formated text, and executes and embeds any R code seamlessly within the document. Whether or not it gets converted to an html or pdf document will depend on the specification you made in your YAML (again, I recommend knitting to HTML).
The rendered document will appear in a pop-up window. If your prefer (as I do) to view this document in the Viewer pane, click the gear icon (located beside the Knit button) and select Preview in Viewer Pane.

You can embedded R code within an Rmd document in two ways:
Then general syntax for inline R code is: `r expression`, where r tells RStudio that expression should be treated and executed as R code. For example, For example: `r 2 + 4` gets rendered to: 6. Notice how this inline code does not appear on its own line and embeds seamlessly within our text.
You can embed an R code in a “chunk”. For example:
gets rendered to
speed dist
Min. : 4.0 Min. : 2.00
1st Qu.:12.0 1st Qu.: 26.00
Median :15.0 Median : 36.00
Mean :15.4 Mean : 42.98
3rd Qu.:19.0 3rd Qu.: 56.00
Max. :25.0 Max. :120.00 Unlike inline R code, chunks appear in a block that is distinct from the written text. Notice how the R input is highlighted in a dark grey box, while the output is boxed with a white background. There is a number of code chunk options you can specify inside the {} (and after the r). For instance, to suppress the R input, you can specify echo = FALSE in the code chunk options. If you wish to suppress the R output, you can include results="hide".
Example 1: suppress R input
gets rendered to
[1] 7Example 2: suppress R output
gets rendered to
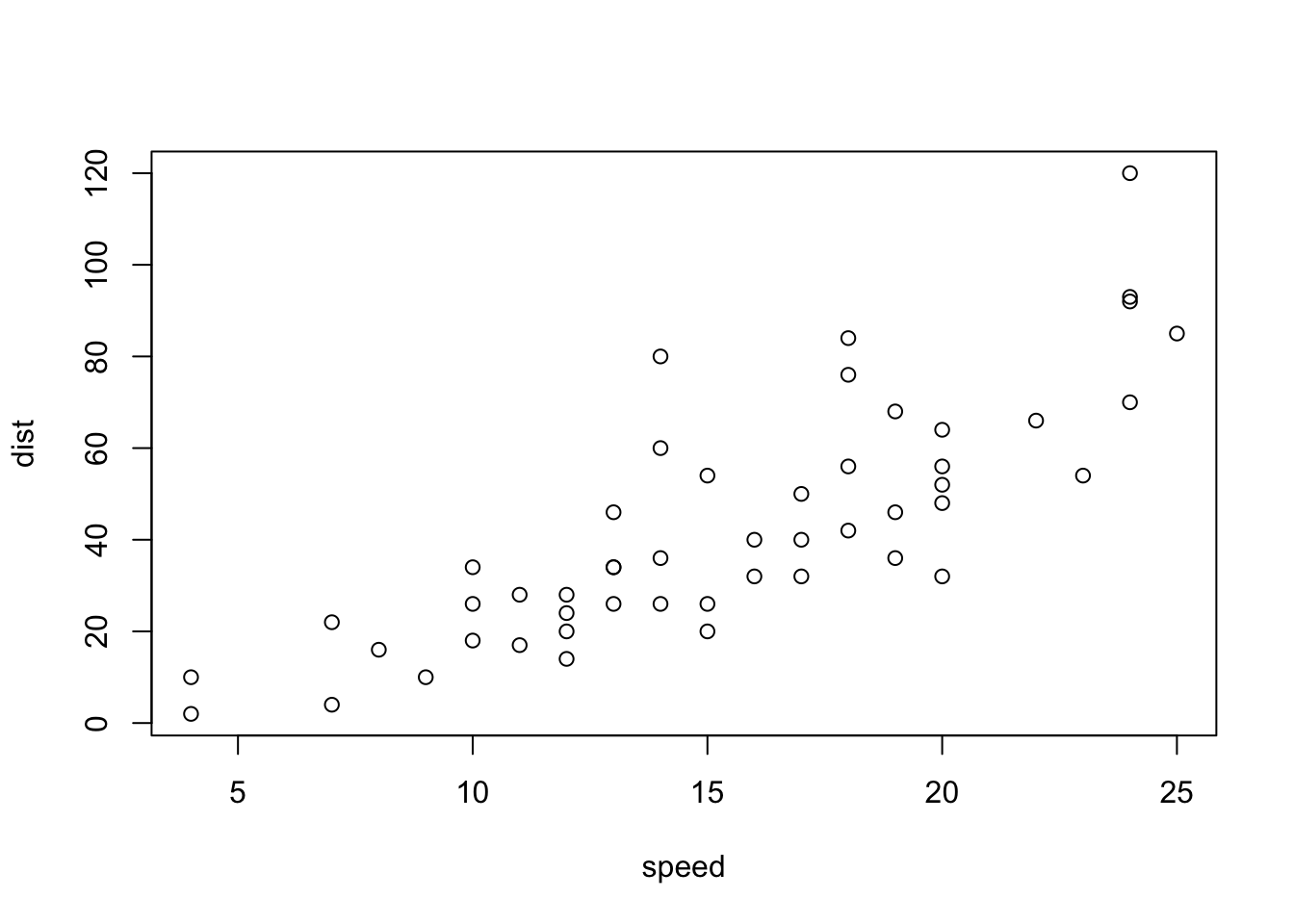
Example 3 embedding plots:
attach(cars) # creates the variables speed and dist from the columns of cars
plot(speed, dist, xlab= "Speed (mph)", ylab = "Distance (m)",
main = "Cars Scatterplot")
If you so desire, you can alter some of the default settings for these R generate plots, eg. resize, alignment, captions. (see code chunk options). For example, we could scale the image to take up 40% of the page width using:
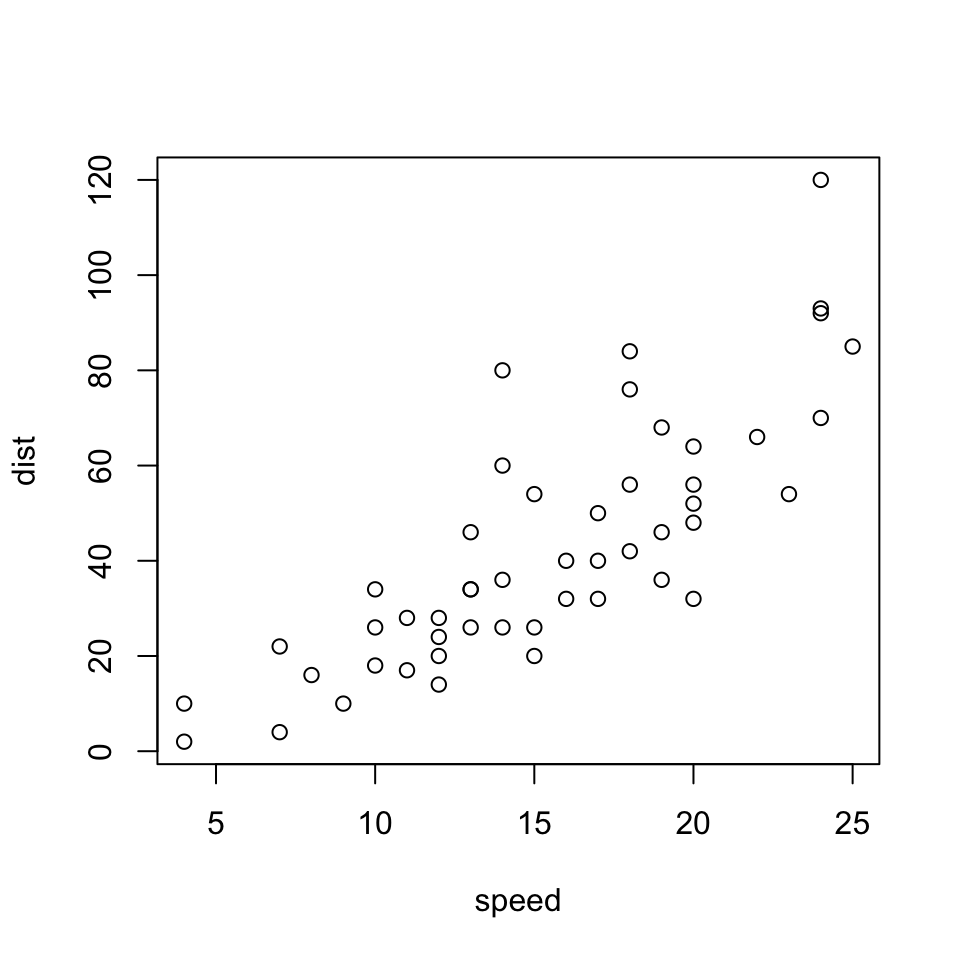
We can also resize the image with fig.height and fig.width (the defaults for both is 7 inches):
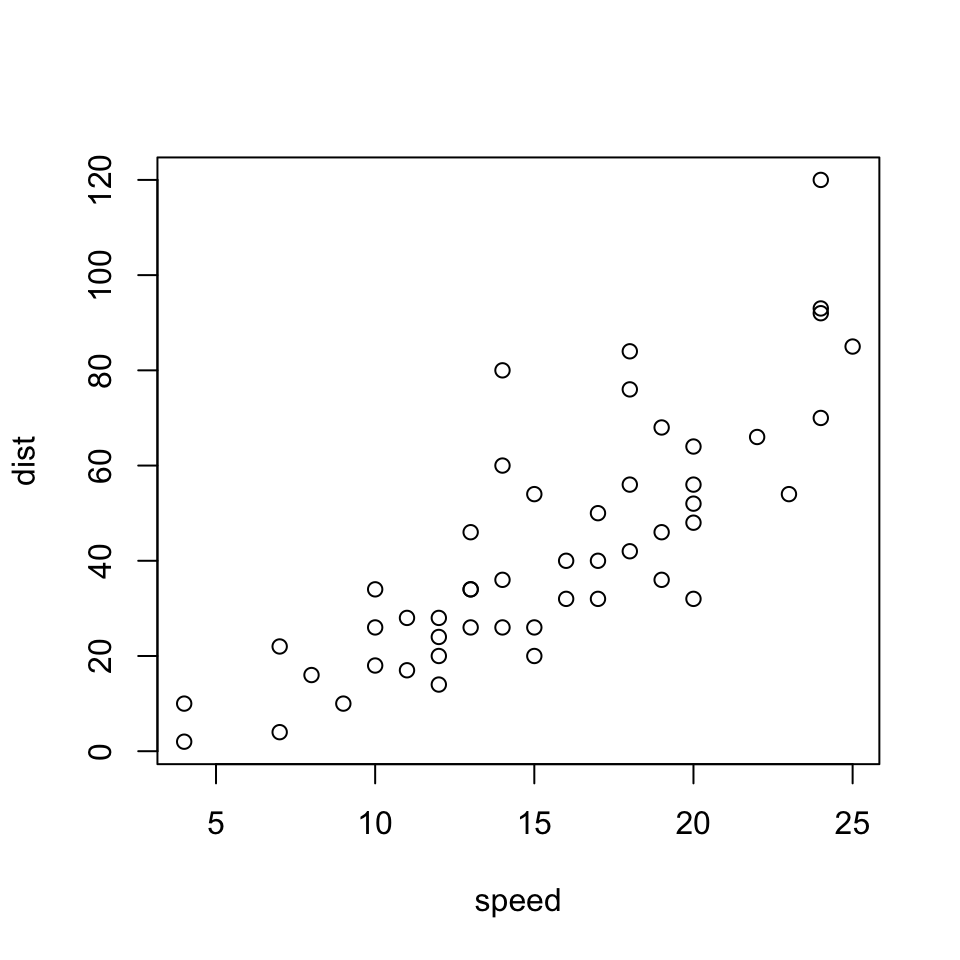
Below I’ve done a combination of both scaling and resizing and centered the image:
```{r echo=FALSE, out.width='40%', fig.height=5, fig.width=5, fig.align="center"}
plot(speed, dist)
```
If you are working within an R Markdown document your working directory is the folder that contains the Rmd file. For instance, a recommended folder system might look something like this:
data311/
|-- labs/
| |-- Lab00.Rmd
| |-- practice00.Rmd
| |-- datamatrix.csv
| |-- data/
| | |-- salaries.csv
|-- assignments/
| |-- template.Rmd
| |-- example.csv
|-- data/
| |-- energydata.csvIn the practice00.Rmd file, I could import datamatrix.csv without a path since it is in our working directory. More generally, when you refer to external files you will need to specify the path relative to the working directory of the Rmd file.
Notably, you cannot change the working directory within a R markdown file using setwd(). You can read more on the subject in Ch 16.6 or the R markdown Cookbook book.
Create an Rmarkdown document (directions) and save it to a meaningful location on your computer. Call this document lab00_x.Rmd where x is replaced by your student number.
Edit the YAML of this document to have the following information:
Delete any auto-populated text within the document (apart from the YAML) an create the following:
echo=FALSE as an R chunk option to hide the R input from view):
st_name your full namest_num your student number Test your use of inline R code by referencing the variables you just created in the following sentence:Hello! My name is <insert name here>. My student number is <insert student number here>.
Display the current version of R your are running using the command version$version.string. If you are not running the current version (see https://www.r-project.org/). Please update your R and recompile this Rmd file.
Read in some data.
datamatrix.csv file and save it to the same folder as your lab00_x.Rmd file.mat. (Ensure that you are also able to do this step using the URL: https://irene.vrbik.ok.ubc.ca/data/datamatrix.csv)mat.Do some calculations. Be sure to set echo = TRUE so that your calculations are printed to screen and clearly number/label/indicate (either using comments in the R chunks or plain text) what you are calculating.
sleep columnfemale entries in the gender columnCreate a scatterplot. Using the mat data frame create a scatterplot with the folling attributes:
sleep on the \(x\)-axis and dread on the \(y\)-axisAdditional resources for R markdown
3. While you may not understand all of the syntax used in this document, you can open the Lab0.Rmd file to see all the underlying syntax that went into building this lab↩︎