Data 311: Machine Learning
Lecture 10: Clustering
University of British Columbia Okanagan
Recall
The goal in supervised setting is to predict \(Y\) (either continuous or categorical) using \(p\) features \(X_1, X_2, \dots, X_p\) measured on \(n\) observations.
In unsupervised learning we do not have a response variable \(Y\) so the goal is to discover interesting things about the measurements on \(X_1, X_2, \dots X_p\)
Goal of Unsupervised Learning
The goal is to discover interesting things about the measurements:
e.g. is there an informative way to visualize the data?
e.g. can we discover subgroups among the variables or among the observations?
e.g. can we discover interesting patterns, relationships, or associations among items or variables in a dataset?
Practical Applications
Techniques for unsupervised learning are of growing importance in a number of fields. For instance:
- search for subgroups among breast cancer patients in order to gain a better understanding of the disease
- understand customer buying behavior to identify groups of shoppers with similar browsing and purchase histories (for targeted adds)
- representing a high-dimensional data set (e.g. gene expression) in smaller dimensions
Practical Applications
Techniques for unsupervised learning are of growing importance in a number of fields. For instance:
- search for subgroups among breast cancer patients in order to gain a better understanding of the disease (clustering)
- representing a high-dimensional data set (e.g. gene expression) in smaller dimensions (dimensionality reduction)
- understand customer buying behavior to identify groups of shoppers with similar browsing and purchase histories (for targeted adds) (association)
Challenges and Advantages of Unsupervised Learning
Challenge: Unsupervised learning is more subjective than supervised learning, as there is no simple goal for the analysis, such as prediction of a response.
Advantage It is often easier to obtain unlabeled data—from a lab instrument or a computer—than labeled data, which can require human intervention.
Types of Unsupervised Learning
Unsupervised learning is utilized for three main tasks [1]:
- clustering
- dimensionality reduction
- association
This lecture will focus on two clustering algorithms: Hierarchical Clustering and K-means Clustering
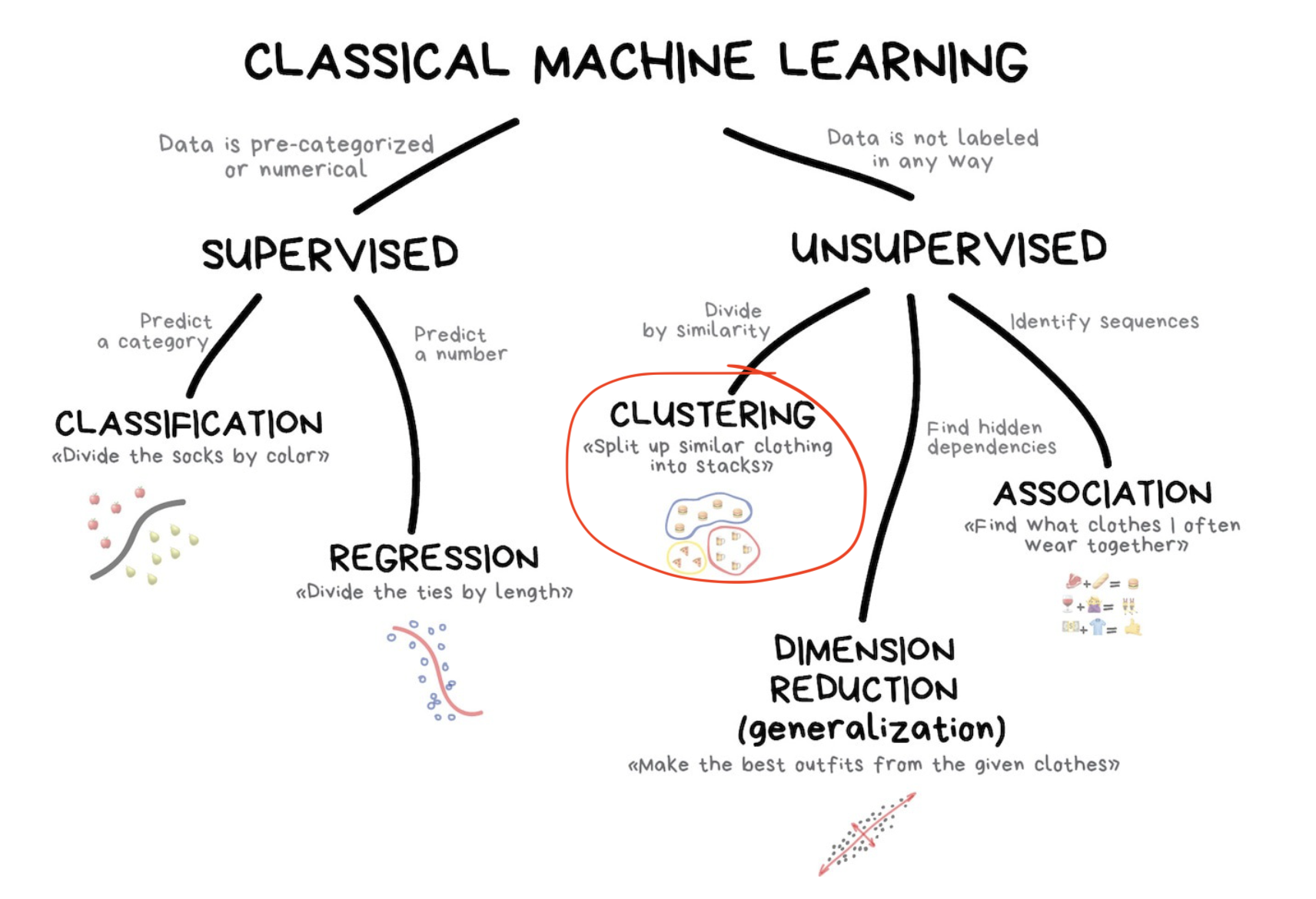
Clustering
The goal of clustering is to find groups (or clusters) such that all observations are
- more similar to observations inside their group and
- more dissimilar to observations in other groups.
Types of clustering algorithms include:
- Hierarchical
- Partitioning (e.g. k-means “hard” clustering)
- Probabilistic Clustering (e.g. Finite Mixture models)
Old Faithful
Old Faithful Geyser Data is available in R (see ?faithful) and contains the following information for the Old Faithful geyser in Yellowstone National Park, Wyoming, USA.
-
waitingthe waiting time between eruptions and -
eruptionsthe duration of the eruption
Old Faithful Plot
While there are no true “classes” in this data set, many would argue that there are two natural “clusters”.

Wine
The
winedata from the gclus package comprise 178 Italian wines from three different cultivars that correspond to the wine varietals:Barolo,Grignolino,BarberaRecorded are 13 measurements (eg. alcohol content, malic acid, ash, …)
- Notably, this data set does have a known class!
- However, is often used to test and compare the performance of various classification algorithms.
Wine Data
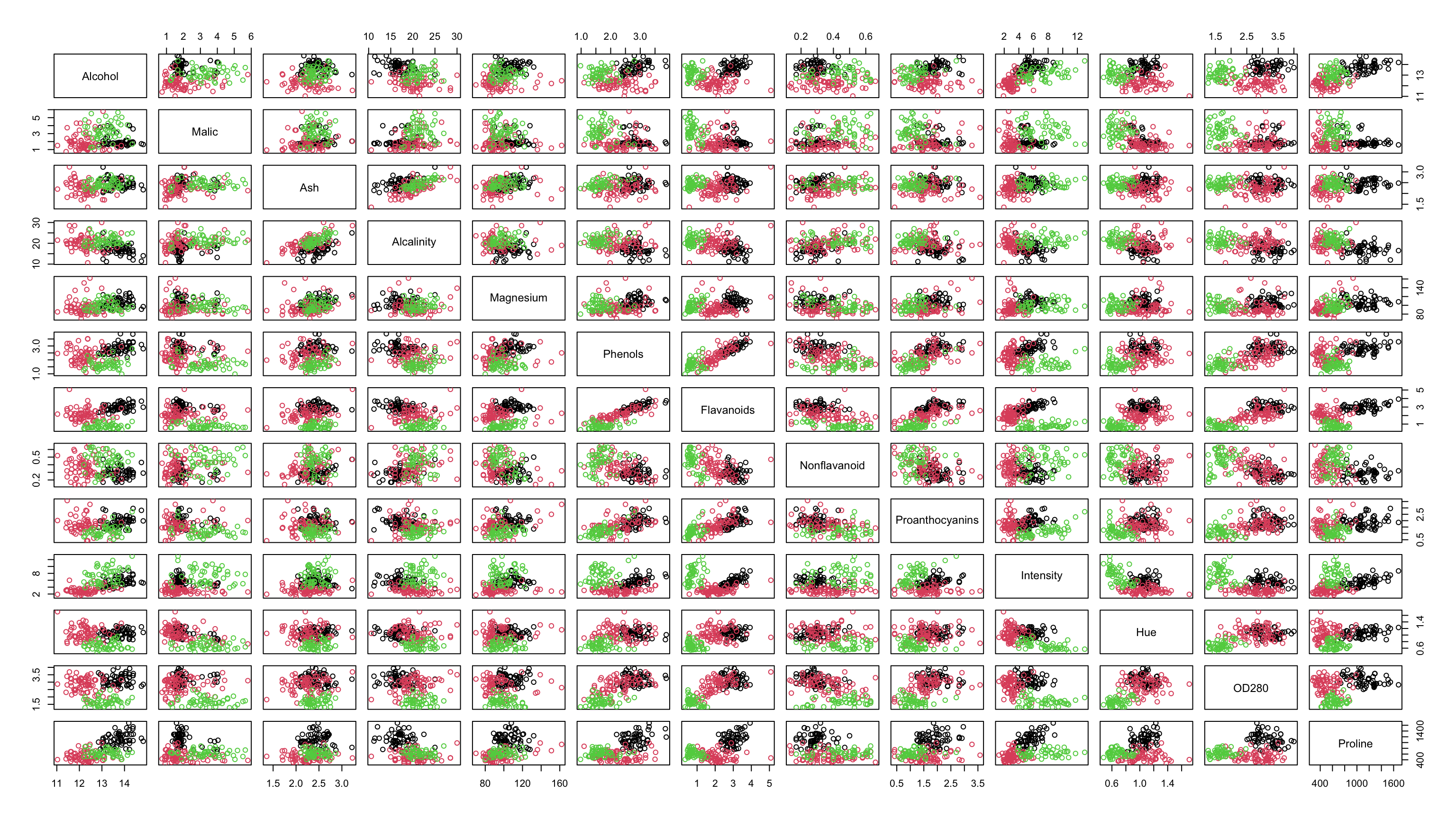
Hierarchical Clustering
Once we have information on the distances between all observations in our data set, we’re ready to come up with ways to group those observations.
Hierarchical clustering (HC), or hierarchical cluster analysis (HCA), is in many ways the most straightforward method for finding groups.
As it’s name suggests, this method builds a hierarchy of clusters.
Types of HC
HC can be categorized in two ways (we’ll focus on 1.):
Agglomerative “bottom-up” approach: each observation starts in its own cluster, and pairs of clusters are merged as one moves up the hierarchy.
Divisive “top-down” approach: all observations start in one cluster, and splits are performed recursively as one moves down the hierarchy.
Agglomerative HC algorithm
Agglomerative HC can be boiled down to the following steps:
Start with all observations in their own groups ( \(n\) unique groups)
Join the two closest observations (now there are \(n-1\) groups)
Recalculate distances (more on this soon)
Repeat 2) and 3) until you are left with only 1 group.
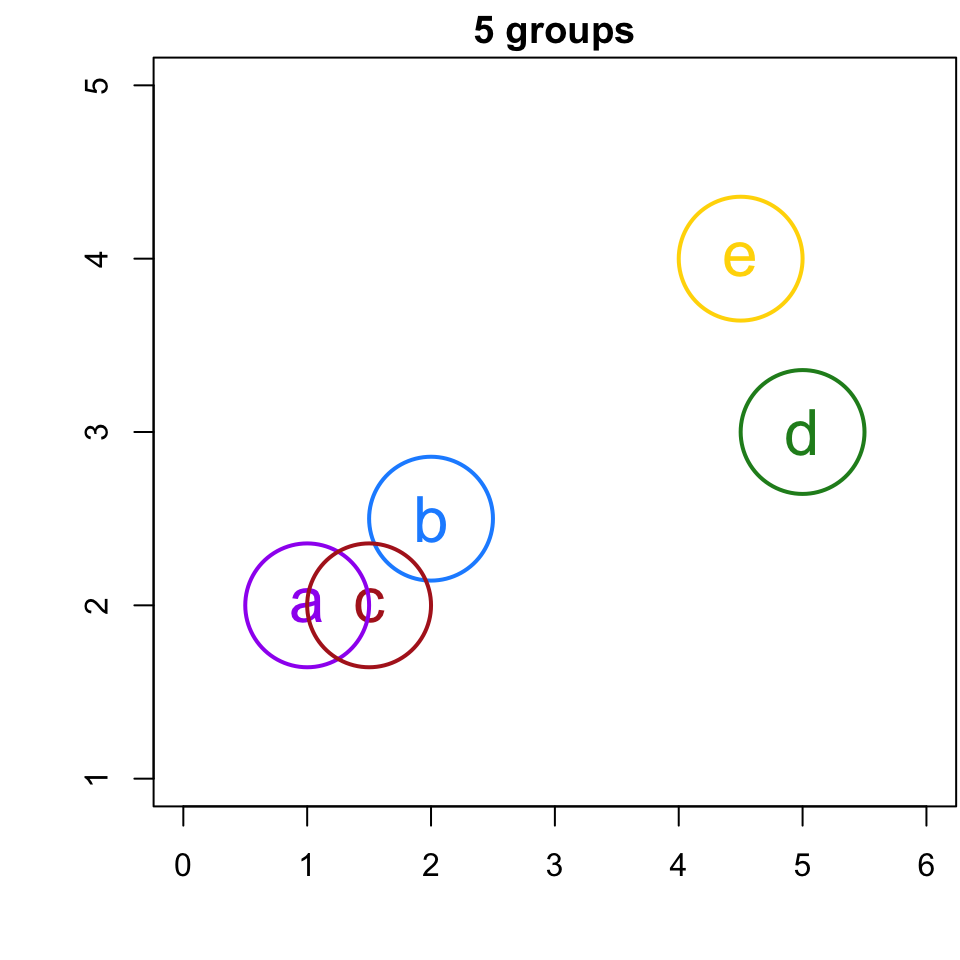
Simple Example
If I were to ask you to group this data, what would you do?
(Again, since we are in the realm of clustering there is no “right” or “wrong” answer).
Now let’s see what HCA produces …
Step 1
Step 2
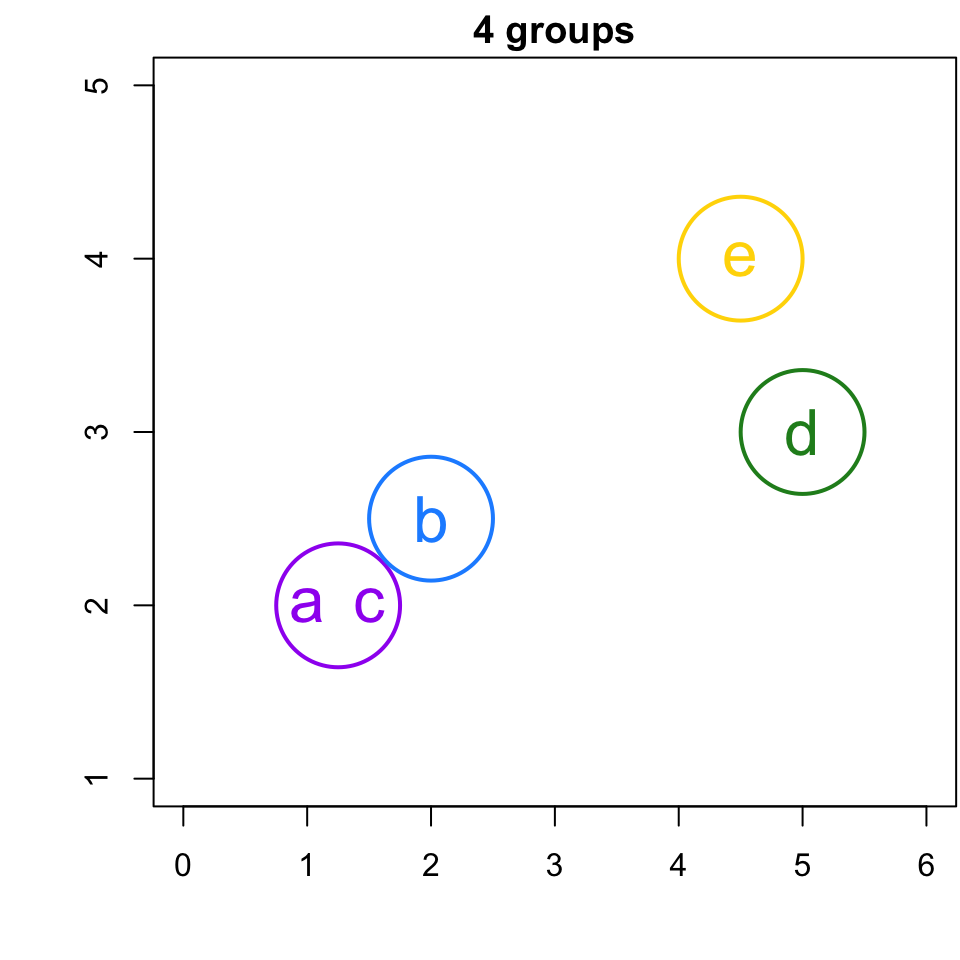
Since a and c are the closest, we join them to a group
| a | b | c | d | e | |
|---|---|---|---|---|---|
| a | 0 | ||||
| b | 1.118 | 0 | |||
| c | 0.5 | 0.707 | 0 | ||
| d | 4.123 | 3.041 | 3.64 | 0 | |
| e | 4.031 | 2.915 | 3.606 | 1.118 | 0 |
Step 3: Recalculating Distances
Now that we’ve joined the two closest observations how do we determine the distance between groups?
For instance, whats the distance between the purple group containing observations a and c and the blue group containing a single observation b? (denote this \(d_{\{ac\}\{b\}}\))
There are 3 common ways of recalculating distances between groups; these are referred to as linkages
Linkages
Single linkage: calculates the minimum distance between two points in each cluster.
Complete linkage: calculates the maximum distance between two points in each cluster.
Average linkage: calculates the average pairwise distance between the observations inside group with those outside.
Centroid Linkage: calculates the distance between the centroids (mean points) of two clusters.
Linkages Example:

- Single linkage: \(d_{\{ac\}\{b\}}=d_{cb}\)
- Complete linkage: \(d_{\{ac\}\{b\}}=d_{ab}\).
- Average linkage: \(d_{\{ac\}\{b\}}=\frac{d_{ab}+d_{cb}}{2}\).
Linkages Example:
| a | b | c | d | e | |
|---|---|---|---|---|---|
| a | 0 | ||||
| b | 1.118 | 0 | |||
| c | 0.5 | 0.707 | 0 | ||
| d | 4.123 | 3.041 | 3.64 | 0 | |
| e | 4.031 | 2.915 | 3.606 | 1.118 | 0 |
- Single linkage: \[d_{\{ac\}\{b\}}=d_{cb} = 0.707\]
- Complete linkage: \[d_{\{ac\}\{b\}}=d_{ab} = 1.118\]
- Average linkage: \[d_{\{ac\}\{b\}}=\frac{d_{ab}+d_{cb}}{2}= 0.9125\]
Distance recalculation
| ac | b | d | e | |
|---|---|---|---|---|
| ac | 0 | |||
| b | 0.707 | 0 | ||
| d | 3.64 | 3.041 | 0 | |
| e | 3.606 | 2.915 | 1.118 | 0 |
- Single linkage: \[d_{\{ac\}\{b\}}=d_{cb} = 0.707\]
Adopting single linkage, our distance matrix would then get updated to something like what you see on the left.
Step 4a

Since group {a,c} and {b} are the closest, we merge them.
| ac | b | d | e | |
|---|---|---|---|---|
| ac | 0 | |||
| b | 0.707 | 0 | ||
| d | 3.64 | 3.041 | 0 | |
| e | 3.606 | 2.915 | 1.118 | 0 |
Step 4b

Recalculate distance:
| abc | d | e | |
|---|---|---|---|
| abc | 0 | ||
| d | 3.041 | 0 | |
| e | 2.915 | 1.118 | 0 |
Step 4c

Since group {e} and {d} are the closest, we merge them.
| abc | d | e | |
|---|---|---|---|
| abc | 0 | ||
| d | 3.041 | 0 | |
| e | 2.915 | 1.118 | 0 |
Step 4d

Recalculate Distance
| abc | de | |
|---|---|---|
| abc | 0 | |
| de | 2.915 | 0 |
Step 4e

Join the two remaining groups: {a,b,c} and {e,d}
| abc | de | |
|---|---|---|
| abc | 0 | |
| de | 2.915 | 0 |
Hierarchy of Clusters
-
What we’ve done is provided a hierarchy of potential solutions to the following non-trivial questions:
- how many groups are in this data?
- what do they look like?
Also, do we really need to go through the algorithm step-by-step to see the process?
Hierarchical Clustering
- We can summarize the algorithm graphically, using what’s called a dendrogram1, i.e. a diagram representing a tree.
On the Y-axis, “Height”, is a measure of closeness of either individual data points or clusters.
Horizontal bars indicate the distance at which two clusters are joined.
Large “jumps” between combining groups signify large distances between groups.
Dendrogram
Dendrogram “jump”

In this case, the dendrogram shows us that there is big difference between cluster {a, b, c} and cluster {d, e} and that the distance within each of these clusters is pretty small.
Determining the Number of Clusters
Dendrograms can also be used to determine how many groups are present in the data.
We can perform a horizontal “cut” in the tree to partitioning the data into clusters.
So generally, we can “cut” at the largest jump in distance, and that will determine the number of groups, as well as each observation’s group membership.
Cut-points (2 group solution)

You can visualize this as an upside-down tree (I have extended the “leaves” of the dendrogram to the the bottom to better explain the cutting process). Where you cut will dictate how many “branches” (i.e. clusters) you end up with.
Dendrogram cut at 1.7
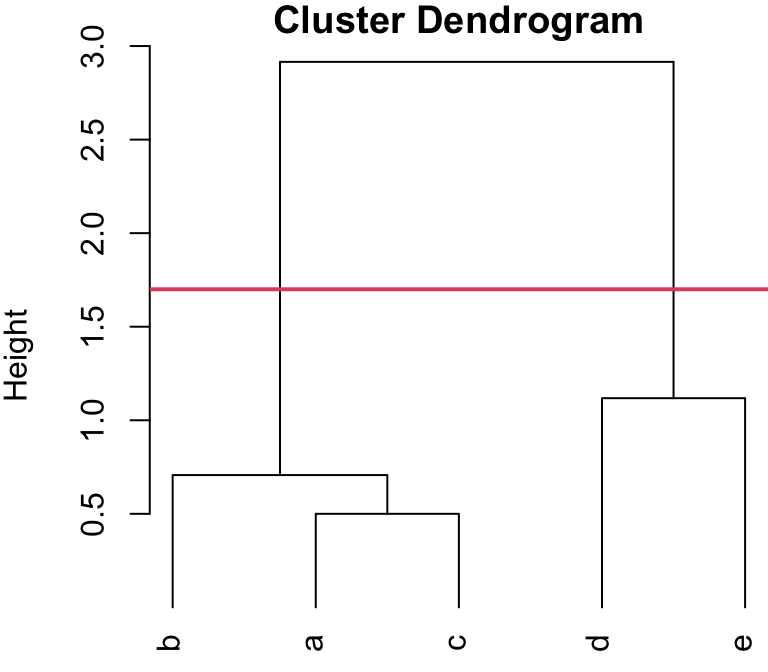
Cutting the dendrogram somewhere along the large “jump” would result in two branches, i.e. clusters
2 group visualization
Which matches our intuition …


We could have cut this tree elsewhere however …
5-group solution
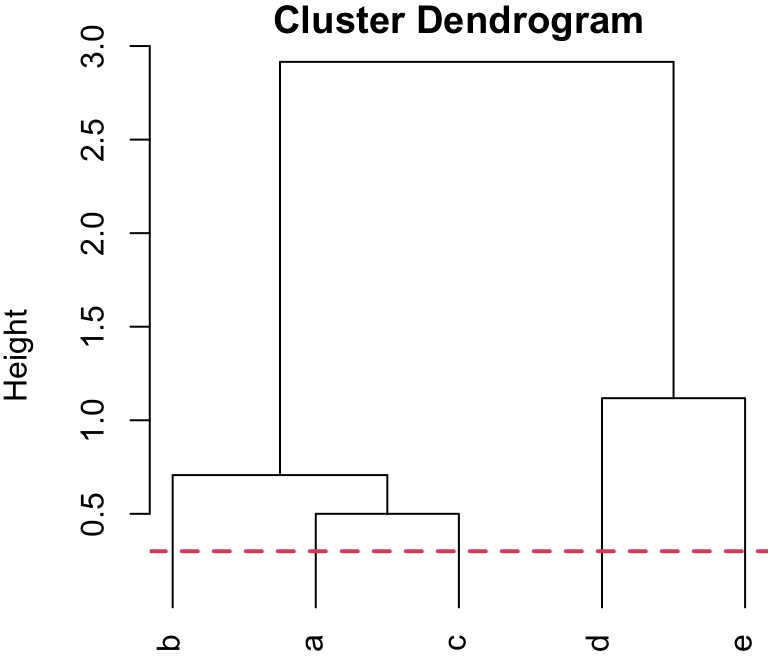
r cutpts[1] or less, would result in 5 singleton clusters, i.e. 5 clusters with only 1 member.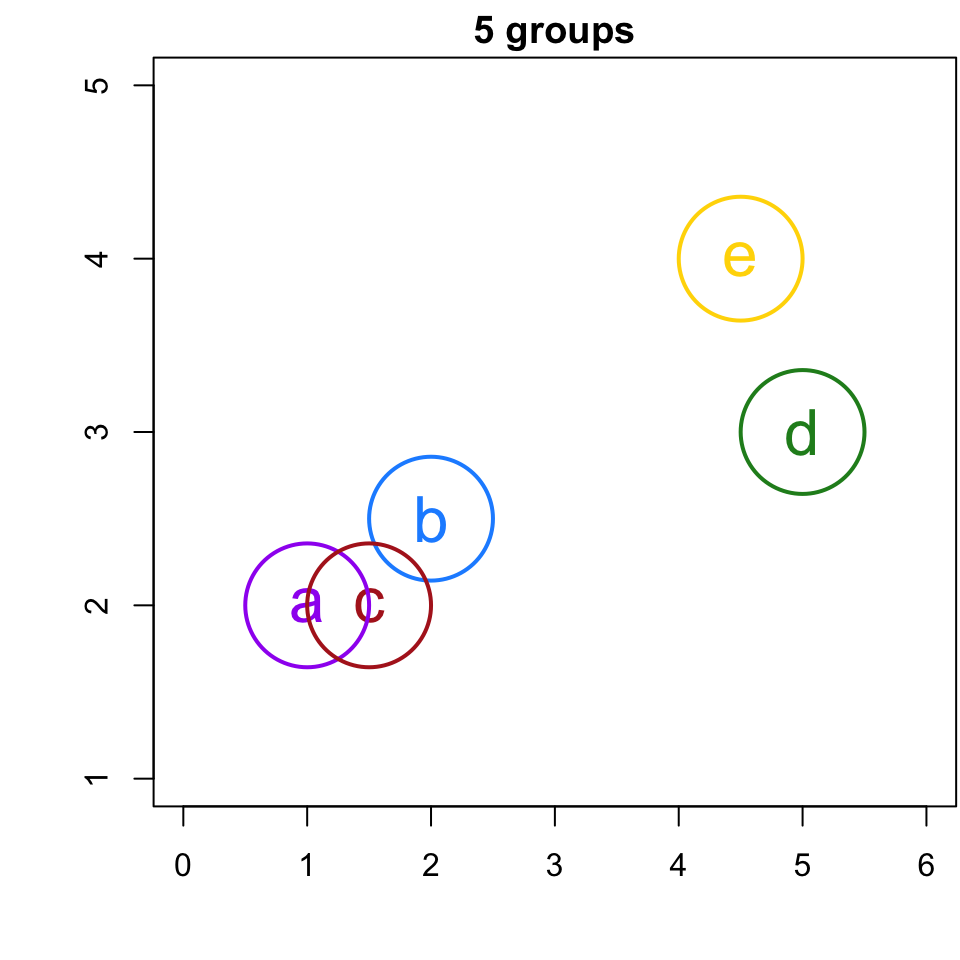
4-group solution
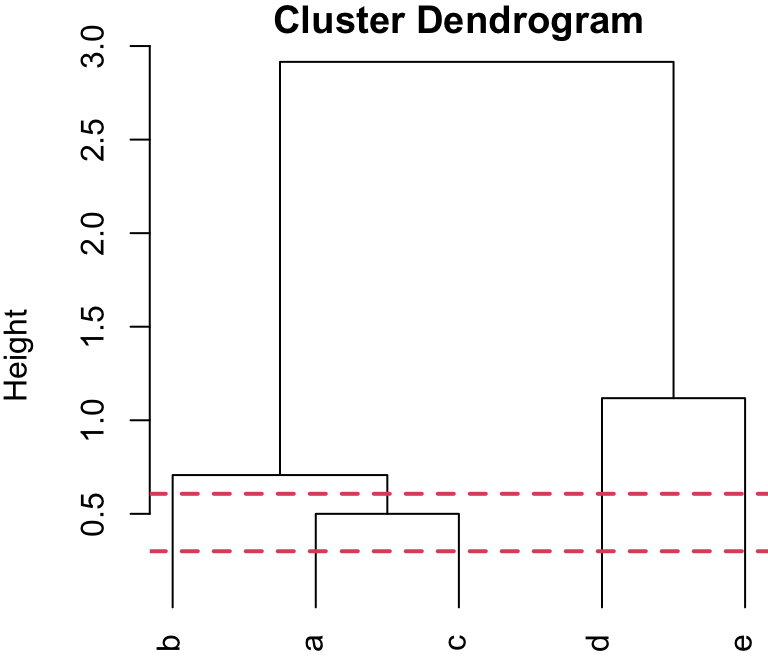
r cutpts[1] and r cutpts[2] , would result in 4 clusters: {b}, {a,c}, {d}, and {e}.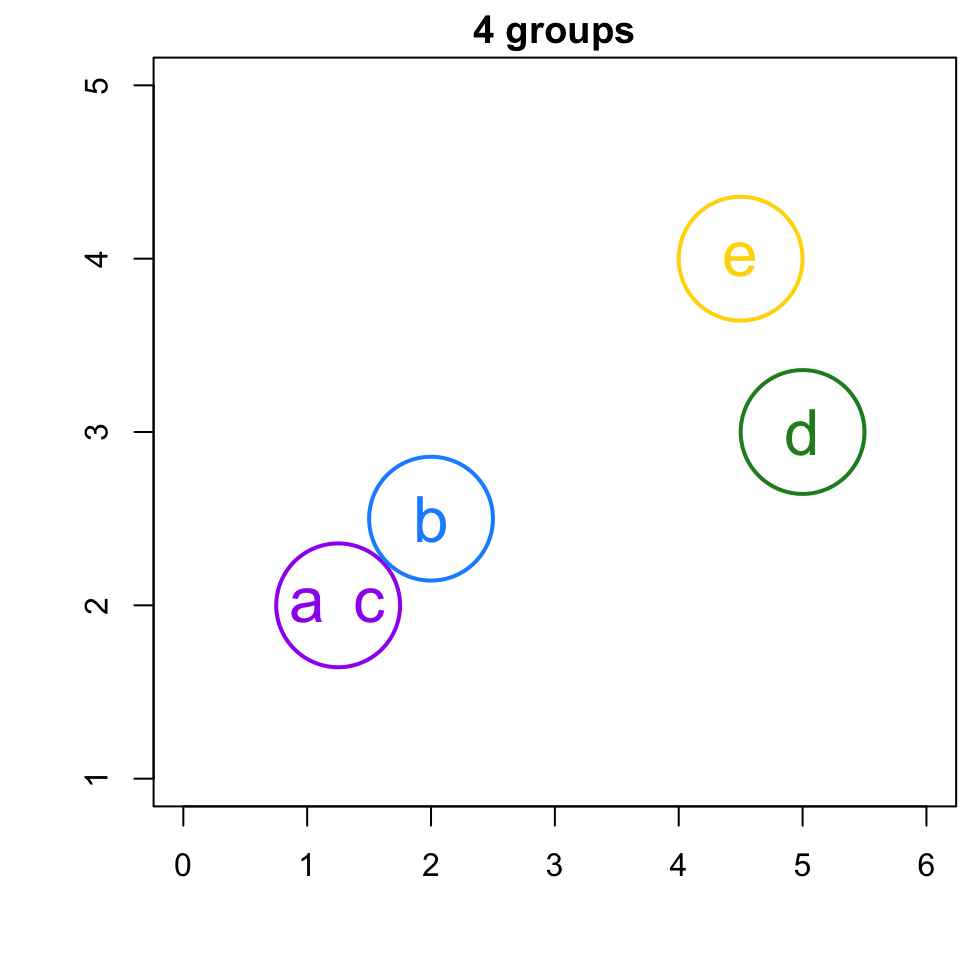
3-group solution

r cutpts[2] and r cutpts[3] , would result in 3 clusters: {b, a,c}, {d}, and {e}.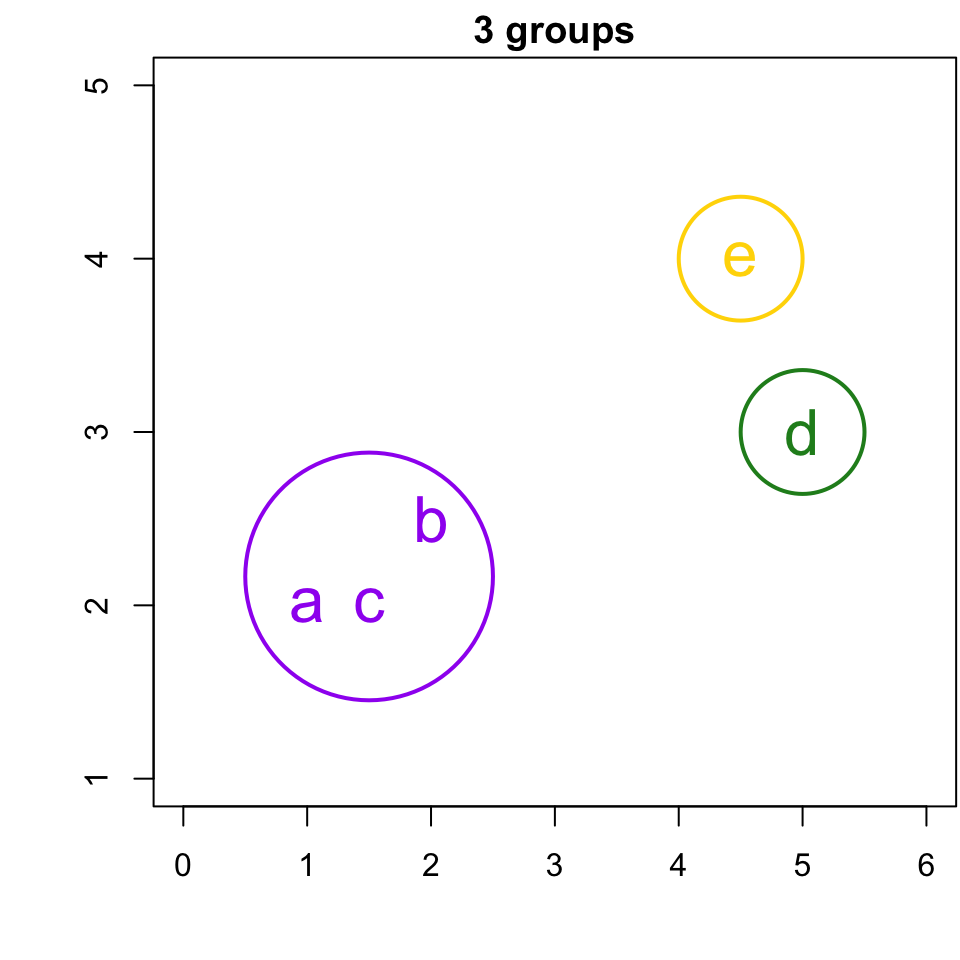
2-group solution
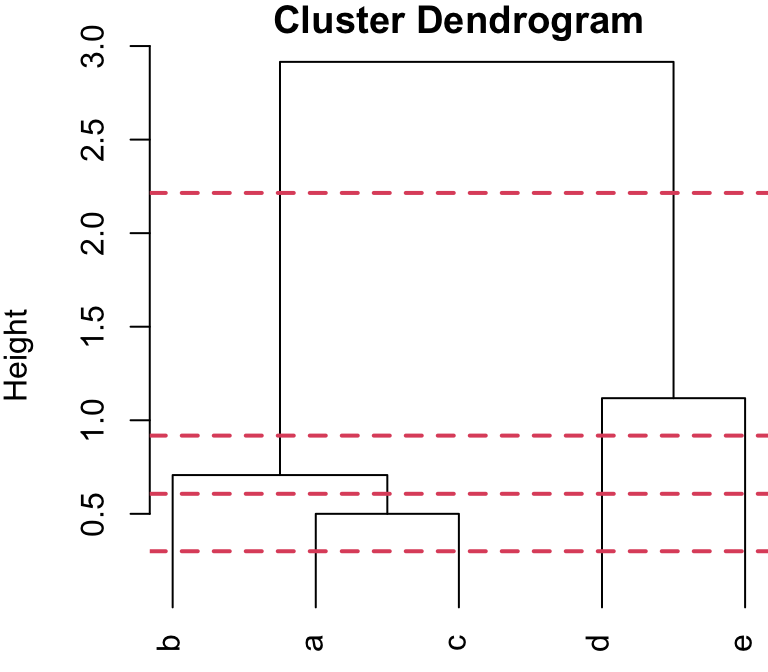
r cutpts[3] and r cutpts[4] , would result in 2 clusters: {b, a,c}, {d, e}.
1-group solution
r cutpts[4] , would result in 1 clusters comprised of all the observations: {a,b, c, d, e}.
HC: Old Faithful
Below is the dendogram on the raw distance matrix (calculated using single linkage) on the Old faithful data


Old Faithful (2 Group Solution)
Here is the two group solution from that model:

Old Faithful (3 Group Solution)
Here is the three group solution from that model:

Old Faithful (4 Group Solution)
Here is the four group solution from that model:

Question
Hmm… These clusters are not ideal.
What do you think might have happened?
- Can you think of something else to try?
HC: Old Faithful (second attempt)
Here is the dendogram on the standardized euclidean distance matrix (using single linkage and the scale function)
hclust() Arguments:
-
d: a dissimilarity structure as produced bydist.
-
method: the agglomeration method to be used. This should be (an unambiguous abbreviation of) one of"ward.D","ward.D2","single","complete","average","mcquitty","median"or"centroid"
HC: Old Faithful (second attempt)
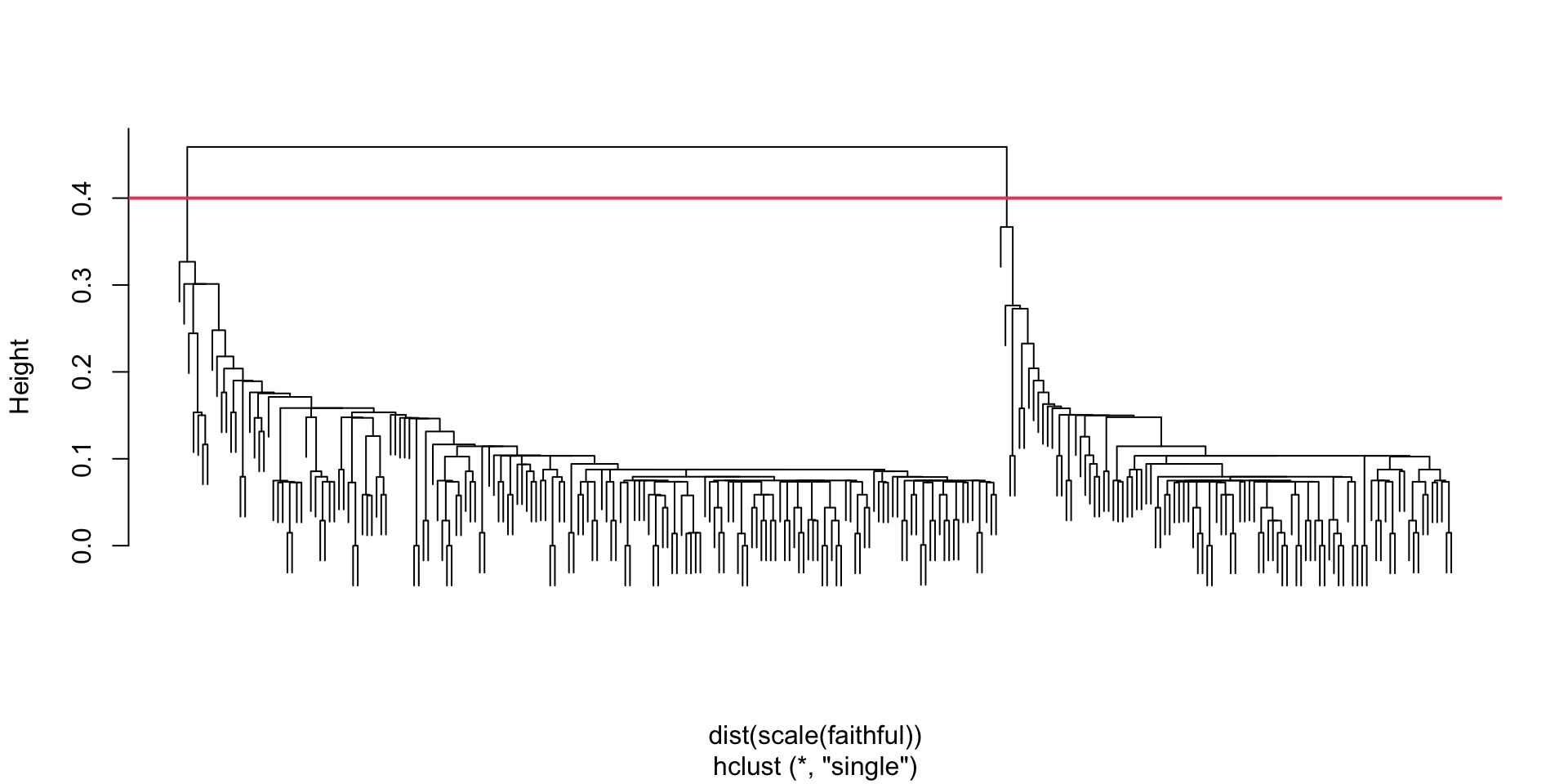
Once we perform HC on the scaled data, we get a two-group solution that much more matches our intuition …
HC: Wine data
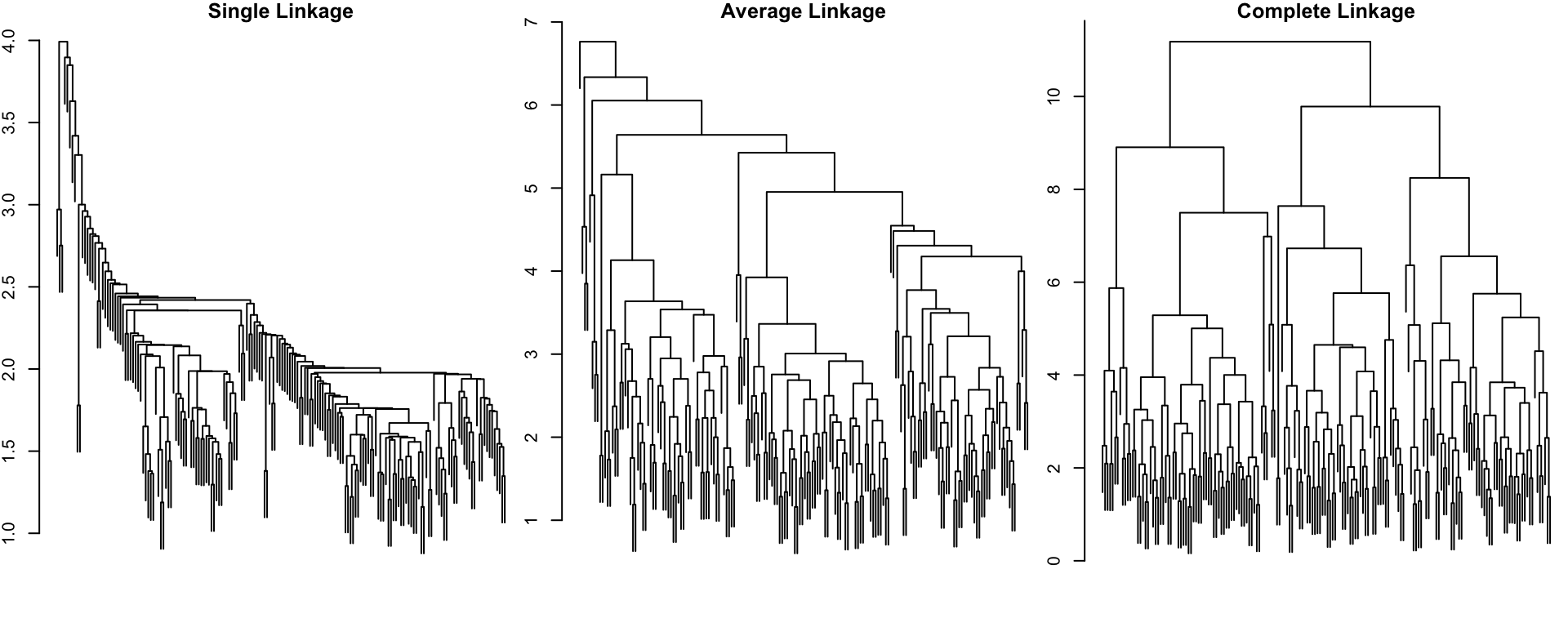
In some cases, the choice of linkage method could make a big difference. Notably, in all cases the data were scaled.
Notes on Linkage
Single-linkage clustering is susceptible to an effect known as “chaining” whereby poorly separated, but distinct clusters are merged at an early stage
Average linkage may lead to a number of singleton clusters (which is generally bad sign)
Complete linkage provides a reasonable partition of the data, suggesting probably 2 or 3 groups. (We’ll come back to this later.)
Comments
Cons
Distance matrices (of size \(n \times n\), symmetric) must be calculated.
For very large samples, this can be time consuming (even for computers).
Results are often sensitive to what distance type and what linkage method are used.
Pro
- easy to implement and understand
K-means Clustering
Next we look at \(k\)-means Clustering.
\(k\)-means clustering is a popular method that requires the user to provide \(k\), the number of groups they are looking for1
In this algorithm, each observation will belong to the cluster whose mean is closest.
After specifying the steps of the algorithm, we will demonstate on the Old Faithful data from last lecture.
Algorithm
- Randomly select \(k\) (the number of groups) points in your data. These will serve as the first centroids1
- Assign all observations to their closest centroid (in terms of Euclidean distance). You now have \(k\) groups.
- Calculate the means of observations from each group; these are your new centroids.
- Repeat 2) and 3) until nothing changes anymore (each loop is called an iteration).
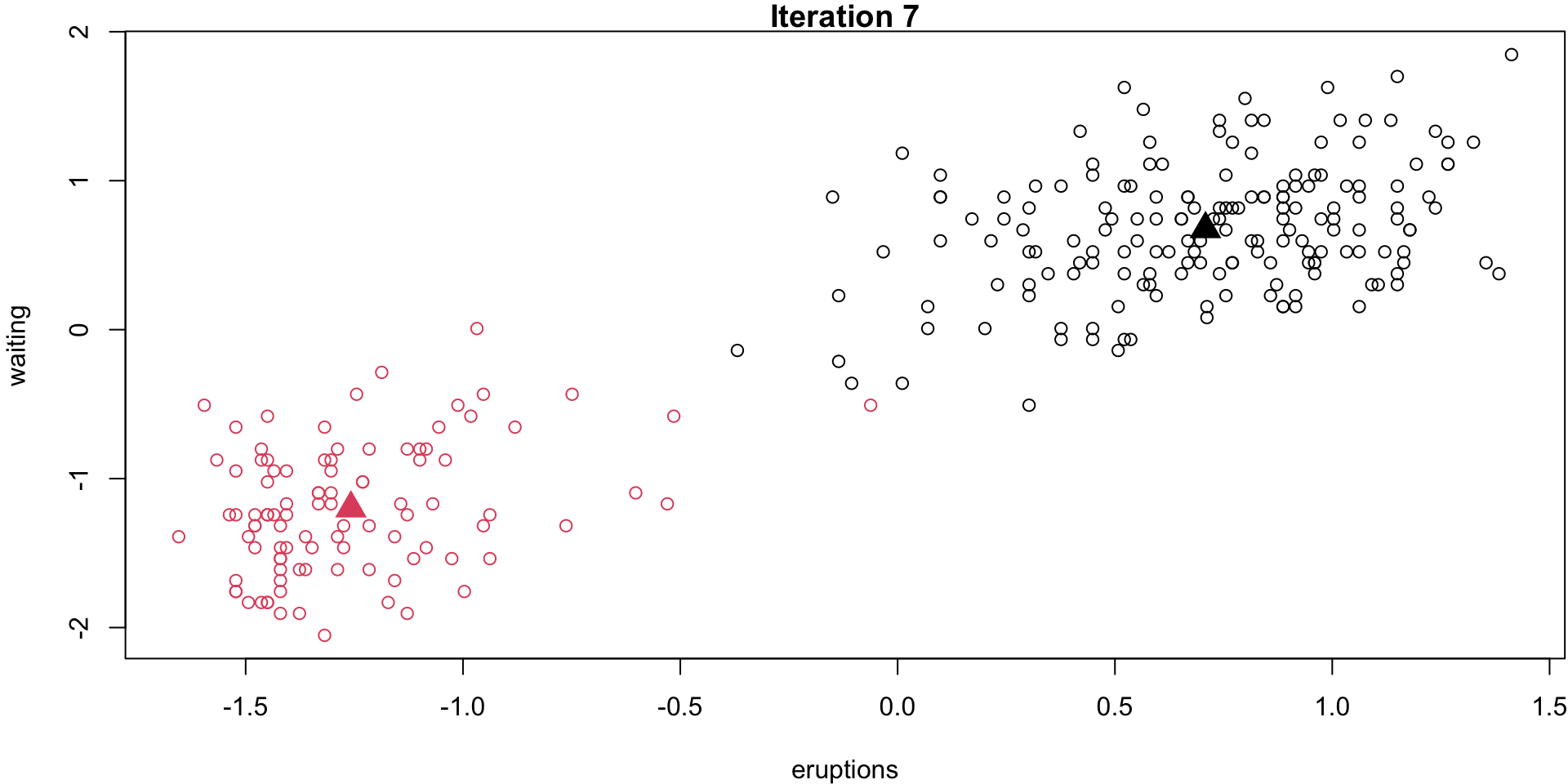
Step 1 (\(k\) = 2)
Randomly select 2 centroid
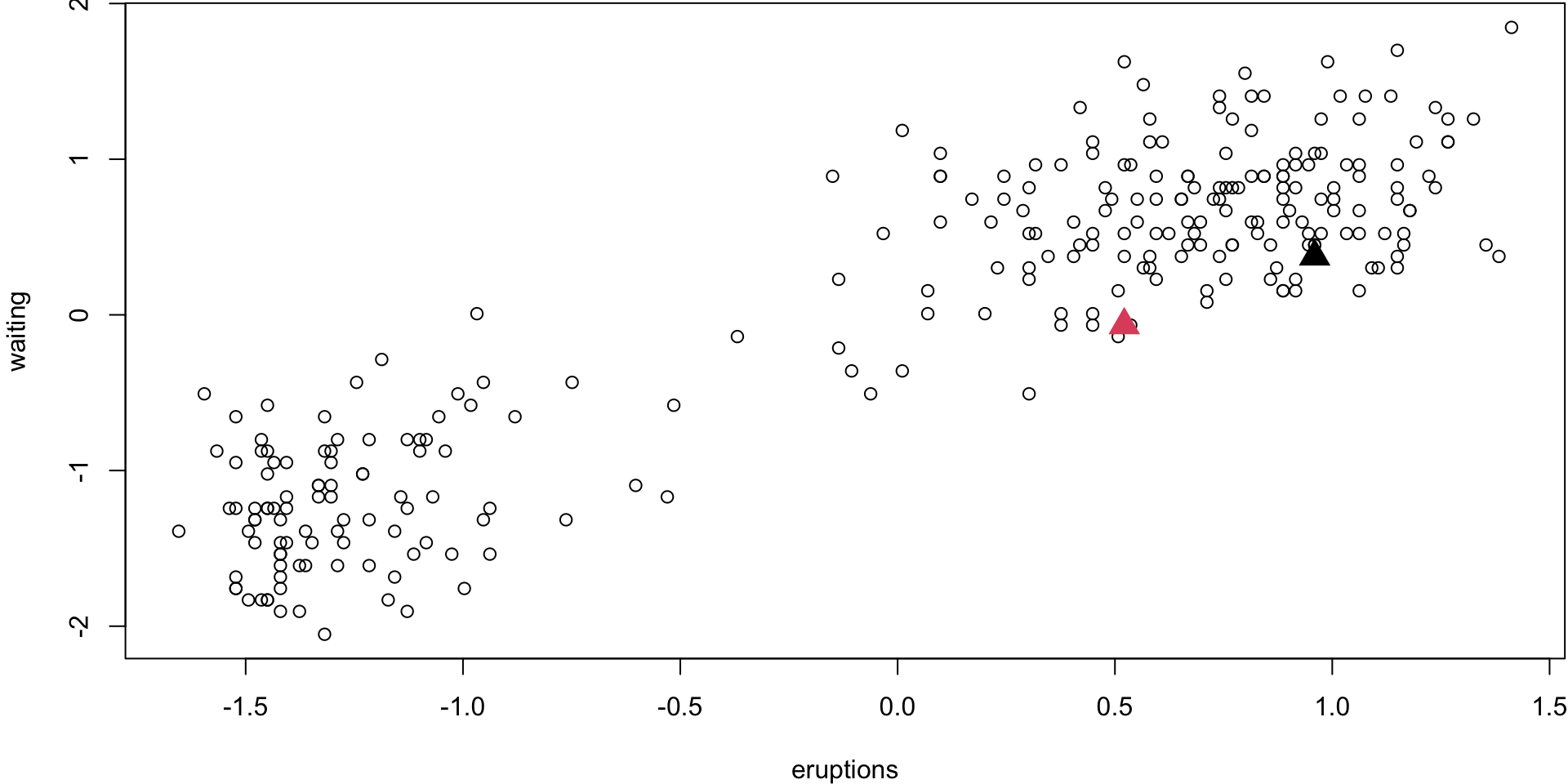
Step 2
Assign obs. to their closest centroid
Code
par(mar = c(4, 4, 0.1, 0.1))
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
plot(x, col=membs)
points(centrs, pch=17, cex=2, col=1:2)
Step 3
Recalculate the means/centroids of each group.

Step 4 (loop)

Code
par(mar = c(4, 4, 1, 0.1))
library(gifski)
count = 2
changing <- TRUE
while(changing){
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
#could write as a double loop, or could utilize other built in functions probably
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
#3) calculate new group centroids
oldcentrs <- centrs #save for convergence check!
for(j in 1:k){
centrs[j,] <- colMeans(x[membs==j, ])
}
plot(x, col=membs, main=paste("Iteration", count))
points(centrs, pch=17, cex=2, col=1:2)
count <- count+1
#4) check for convergence
if(all(centrs==oldcentrs)){
changing <- FALSE
text(0.75, -1, "DONE!", cex=4)
}
}
membs2 <- membs # save 2-group solutionStep 4 (last iteration)
par(mar = c(4, 4, 1, 0.1))
count = 2
changing <- TRUE
while(changing){
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
#could write as a double loop, or could utilize other built in functions probably
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
#3) calculate new group centroids
oldcentrs <- centrs #save for convergence check!
for(j in 1:k){
centrs[j,] <- colMeans(x[membs==j, ])
}
count <- count+1
#4) check for convergence
if(all(centrs==oldcentrs)){
plot(x, col=membs, main=paste("Iteration", count))
points(centrs, pch=17, cex=2, col=1:2)
changing <- FALSE
# text(0.75, -1, "DONE!", cex=4)
}
}Step 4 (last iteration)
Step 1 (\(k\) = 3)
Randomly select 3 observations ( \(k\) =3). These will be your centroids for the 3 groups
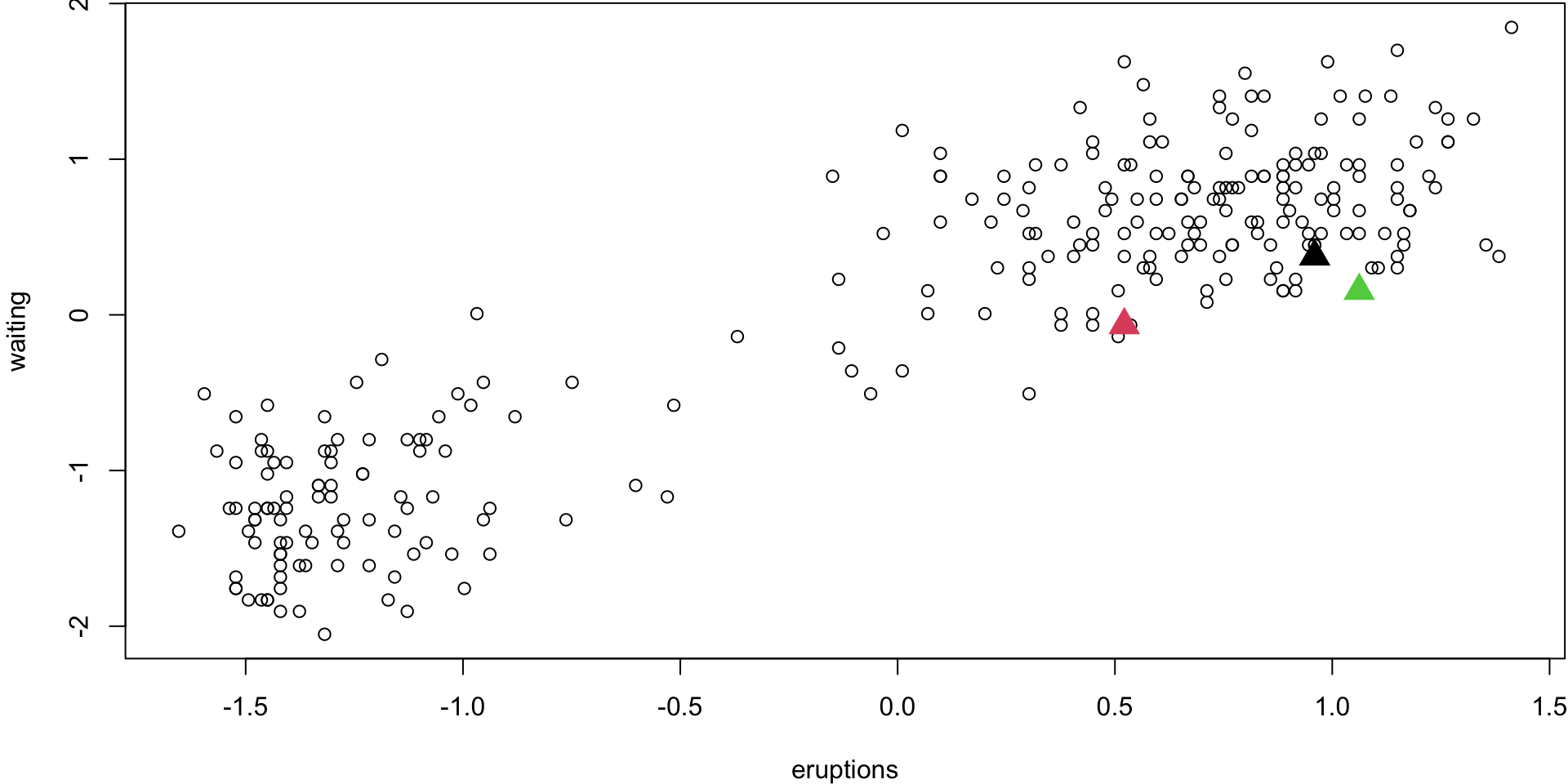
Step 2
Assign obs. to their closest centroid
Code
par(mar = c(4, 4, 0.1, 0.1))
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
plot(x, col=membs)
points(centrs, pch=17, cex=2, col=1:k)
Step 3
Recalculate the means of each group. These are your new centroids.
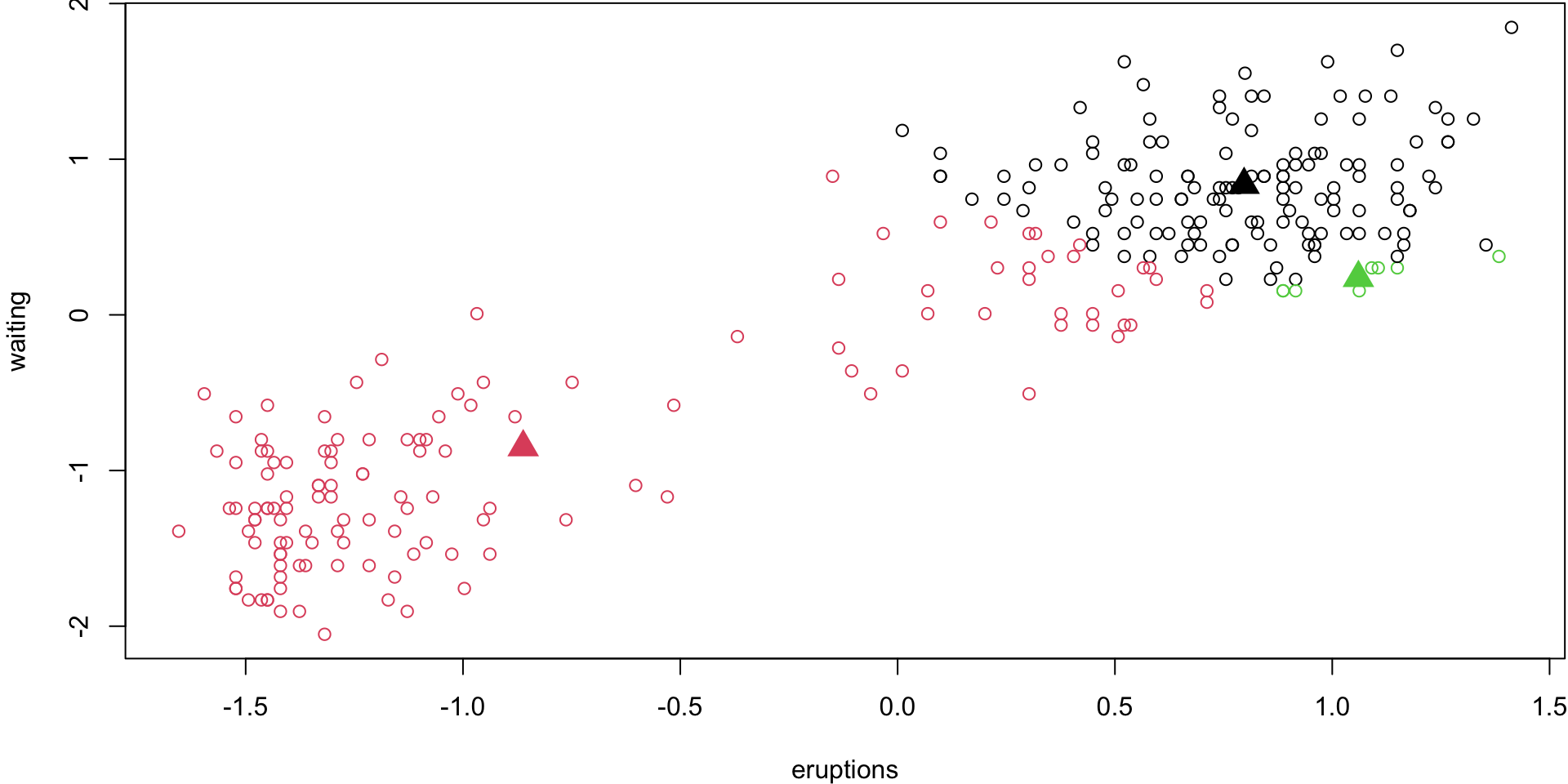
Step 4 (loop)
Repeat 2) and 3) until nothing changes anymore.

Code
par(mar = c(4, 4.1, 1, 0.1))
count = 2
changing <- TRUE
while(changing){
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
#could write as a double loop, or could utilize other built in functions probably
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
#3) calculate new group centroids
oldcentrs <- centrs #save for convergence check!
for(j in 1:k){
centrs[j,] <- colMeans(x[membs==j, ])
}
plot(x, col=membs, main=paste("Iteration", count))
points(centrs, pch=17, cex=2, col=1:k)
count <- count+1
#4) check for convergence
if(all(centrs==oldcentrs)){
changing <- FALSE
text(0.75, -1, "DONE!", cex=4)
}
}
membs3 <- membs # save 3-group solutionStep 4 (last iteration)
par(mar = c(4, 4.1, 1, 0.1))
count = 2
changing <- TRUE
while(changing){
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
#could write as a double loop, or could utilize other built in functions probably
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
#3) calculate new group centroids
oldcentrs <- centrs #save for convergence check!
for(j in 1:k){
centrs[j,] <- colMeans(x[membs==j, ])
}
count <- count+1
#4) check for convergence
if(all(centrs==oldcentrs)){
plot(x, col=membs, main=paste("Iteration", count))
points(centrs, pch=17, cex=2, col=1:k)
changing <- FALSE
# text(0.75, -1, "DONE!", cex=4)
}
}Step 4 (last iteration)
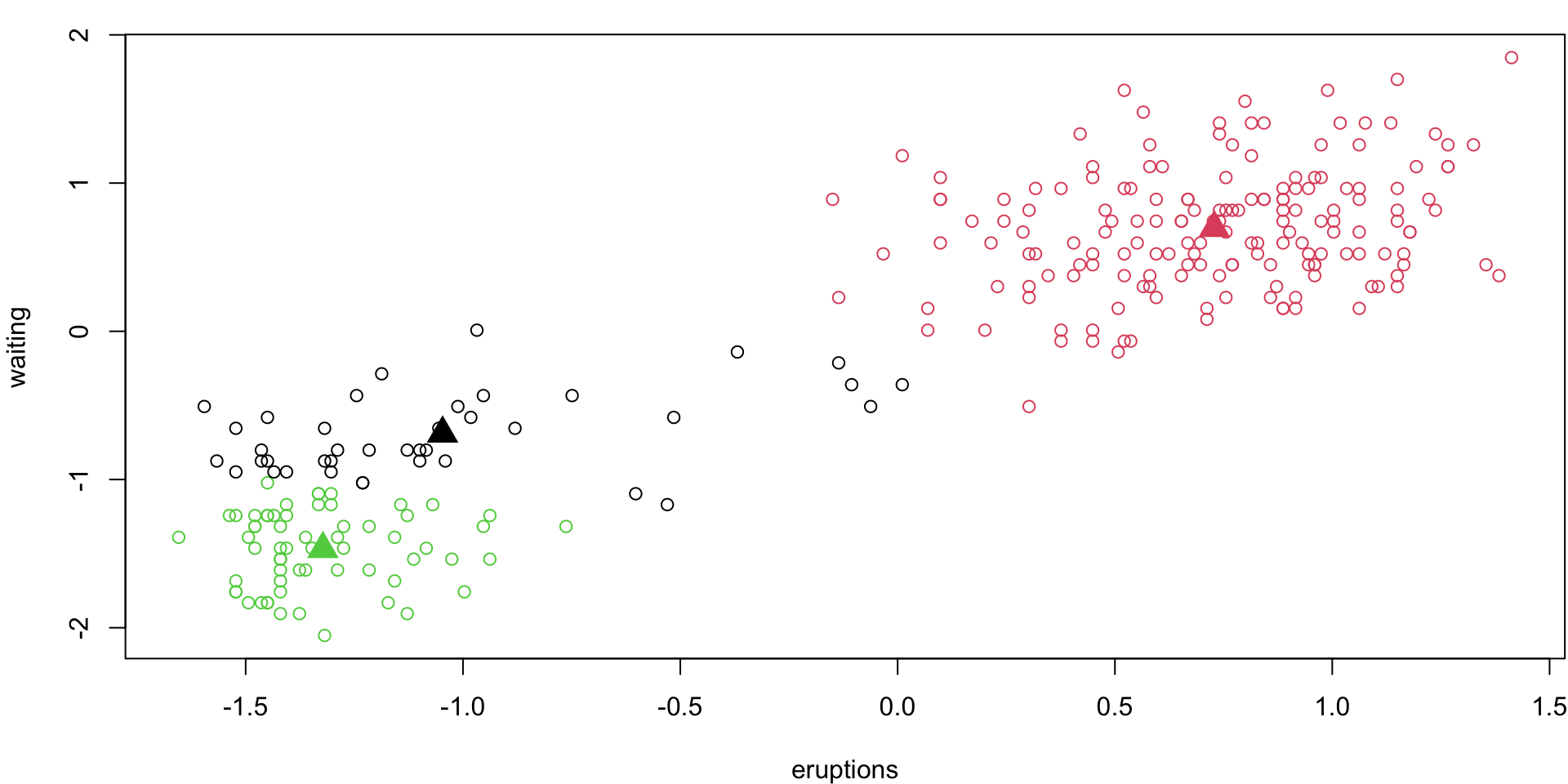
Different 3-group (loop)
Let’s do the 3-group \(k\)-means again:

Different 3-group solution
(Last Iteration)
Code
par(mar = c(4, 4, 1.1, 0.1))
set.seed(1234)
#1 start with k centroids
centrs <- x[sample(1:nrow(x), k),]
#start loop
changing <- TRUE
while(changing){
#2a) calculate distances between all x and centers
dists <- matrix(NA, nrow(x), k)
#could write as a double loop, or could utilize other built in functions probably
for(i in 1:nrow(x)){
for(j in 1:k){
dists[i, j] <- sqrt(sum((x[i,] - centrs[j,])^2))
}
}
#2b) assign group memberships (you probably want to provide some overview of apply functions)
membs <- apply(dists, 1, which.min)
#3) calculate new group centroids
oldcentrs <- centrs #save for convergence check!
for(j in 1:k){
centrs[j,] <- colMeans(x[membs==j, ])
}
# readline(prompt="Press [enter] to continue")
#4) check for convergence
if(all(centrs==oldcentrs)){
plot(x, col=membs)
points(centrs, pch=17, cex=2, col=1:k)
changing <- FALSE
#text(mean(x), -1, "DONE!", cex=4)
}
}
3 group comparison
Depending on our random starting points, we can get very different answers …
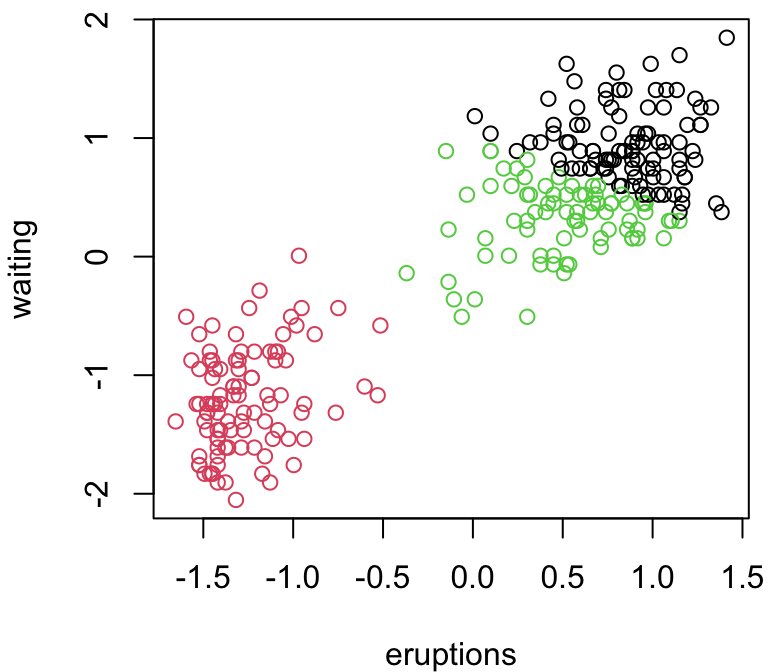
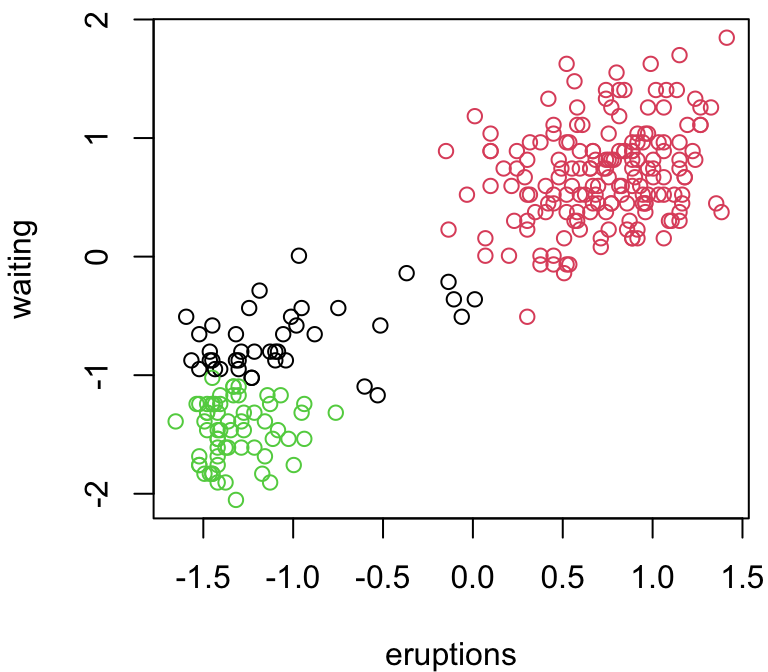
Why does \(k\)-means work?
- Randomly select \(k\) (the number of groups) points in your data. These will serve as the first centroids
- Assign all observations to their closest centroid (in terms of Euclidean distance). You now have \(k\) groups.
- Calculate the means of observations from each group; these are your new centroids.
- Repeat 2) and 3) until nothing changes anymore
2. and 3. are recursively finding the minimum within-group sum of squared distances between points and their centroids.
Comments
Pros
- Computationally efficient, even for large data sets.
- Only \(n \times k\) distance matrices needed
- Relatively straightforward
- Often provides clearer groups than HC.
Cons
- stochastic, i.e. random (as opposed to deterministic)
- can return local optimal rather than global
- not for categorical \(X\)
- Groups will be found no matter what.
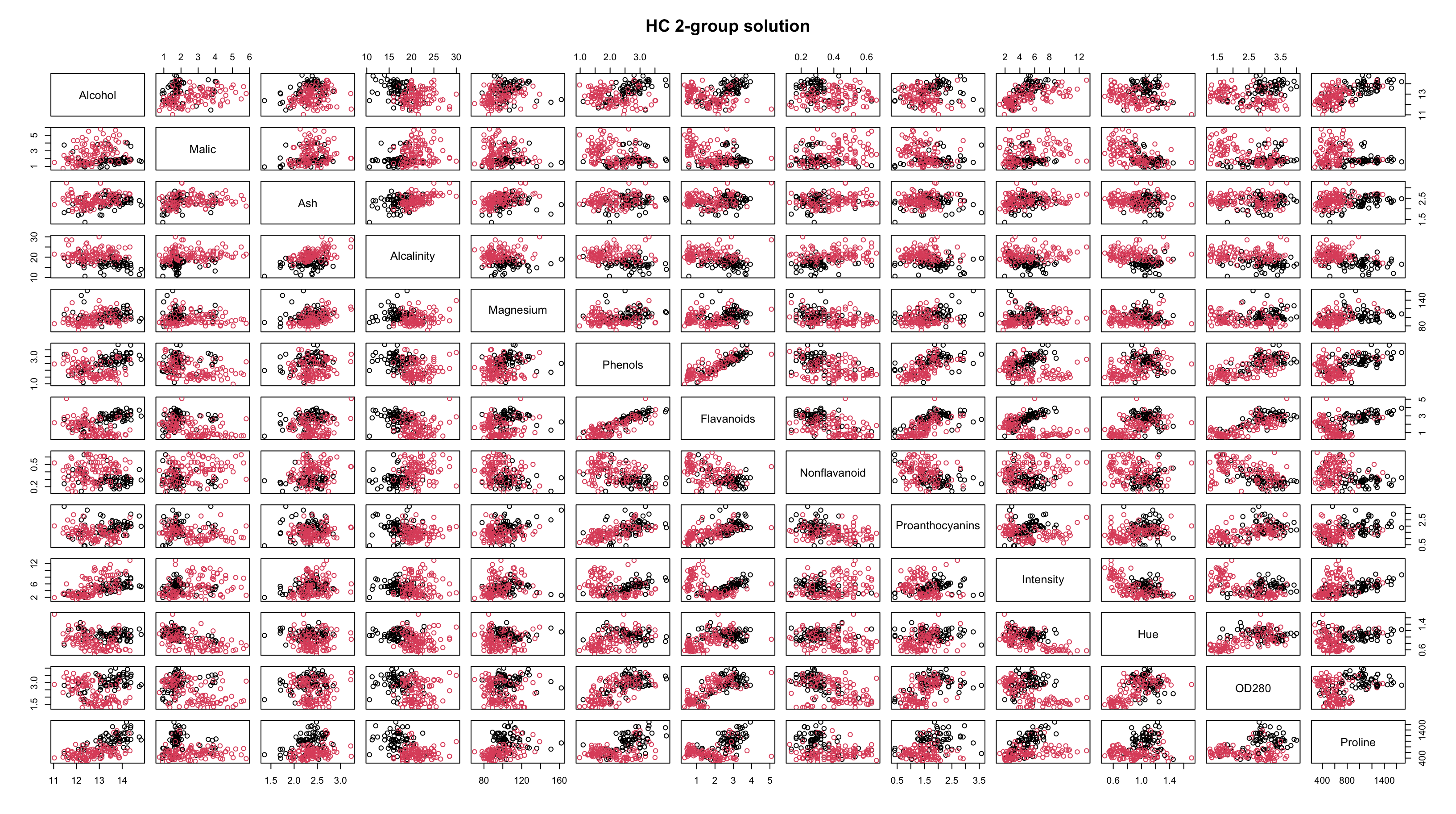
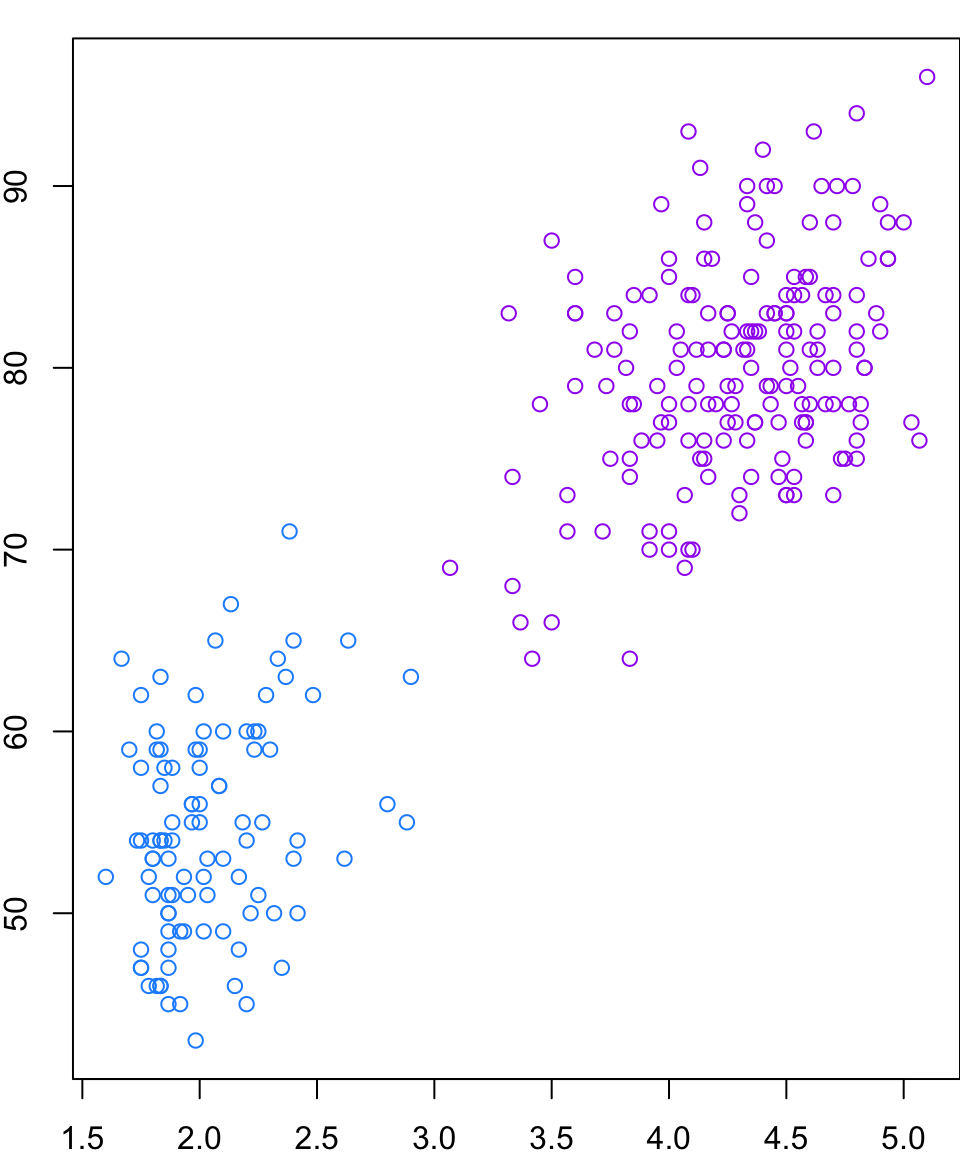
Wine
Let’s return to the
winedata setLet’s apply HC and \(k\)-means and then investigate the results…
Recall that complete linkage appeared to be the most appropriate linkage choice and it suggested 2 or 3 groups.

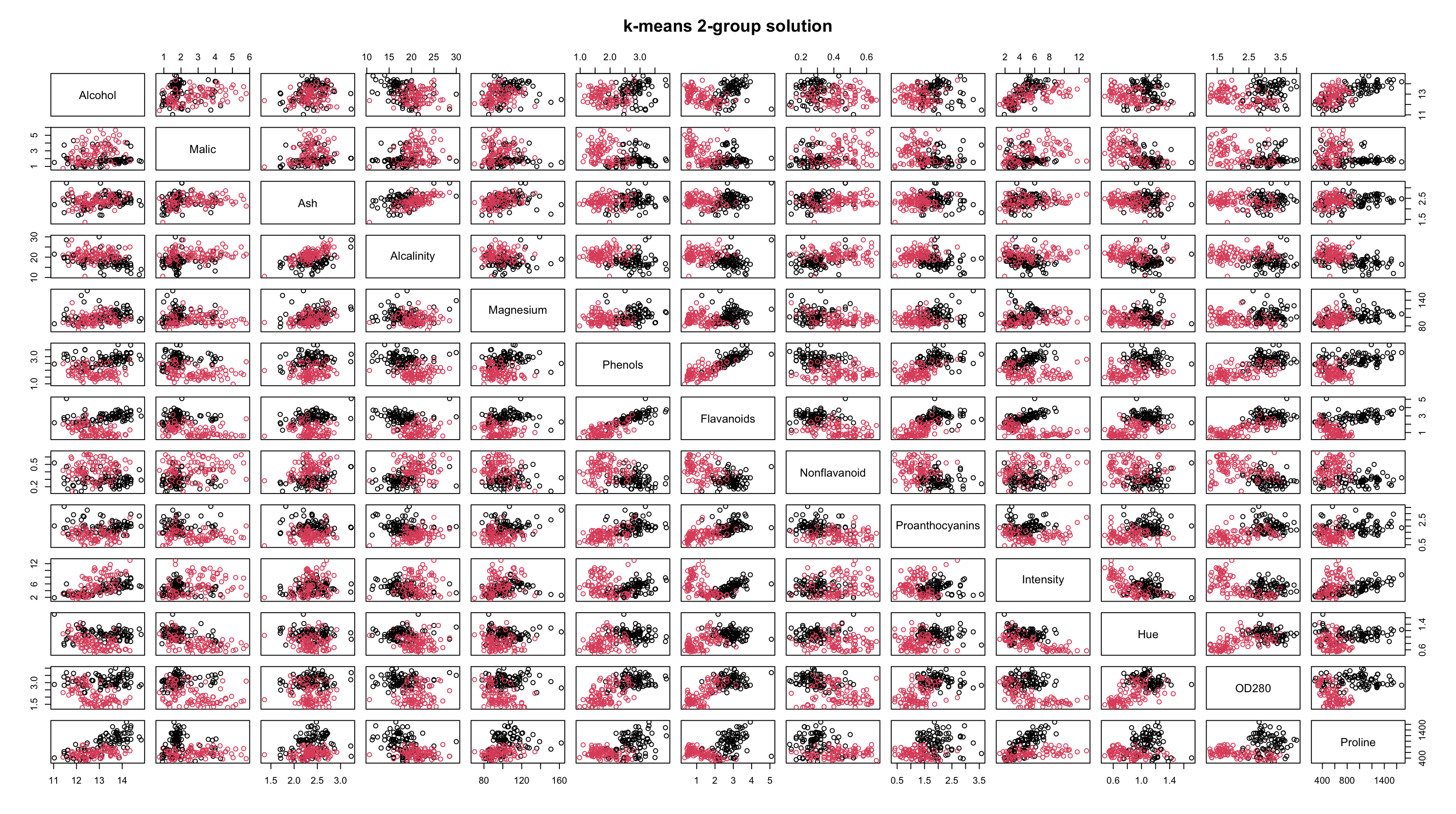
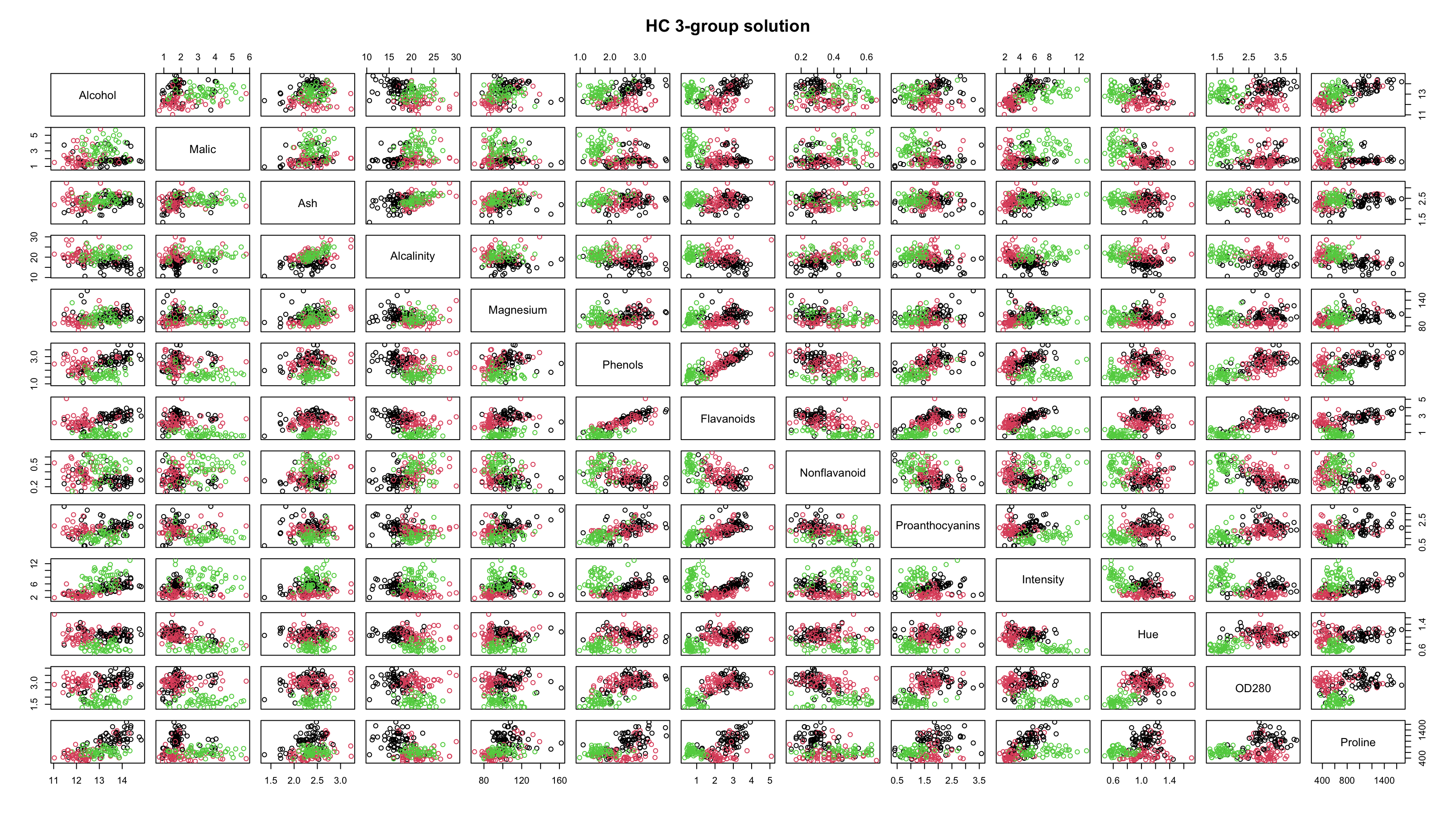

Comparing Results
By looking at the plots, it seems as though the results are pretty similar between \(k\)-means and HC.
The resolution makes it tough to tell, but the solutions are not identical.
-
Both methods are finding most of the group structure, but…
- \(k\)-means only misclassifies 6 of the 178 wines
- whereas HC misclassifies 29 of them
Assessing Results
But remember, we actually have know classes (but hid them in training)
The wine data set among others (e.g.
Iris,crabs) are benchmark data clustering data sets that are used to assess performanceWith the understanding that we wouldn’t be able to do this is a authentic clustering problem, in these scenarios we would be able to build confusions matrices and calcaulte classification error, etc.
Confusion Matrices
HC
Notice how the correctly labeled observations do not necessarily fall on the main diagonal!
Partitioning and Hierarchical
-
Neither HC or \(k\)-means can really be considered “statistical” in the sense of:
- What model are we assuming?
- How can we test whether the model is accurate?
They are basically ad hoc methods that mostly follow from intuition and tend to perform alright.
Eventually, we’ll see clustering via mixture models; the unsupervised equivalent of discriminant analysis.
Dealing with Random Starts
As mentioned, \(k\)-means minimizes the within group sum of squares (WSS); however, it does so locally (not globally)
Furthermore, since it’s initialized randomly, results change from run to run.
Importantly: it’s fast! So we can run it many times, and choose the solution with the smallest within-group sum of squares for those runs.
In fact, there’s a built in argument in R…
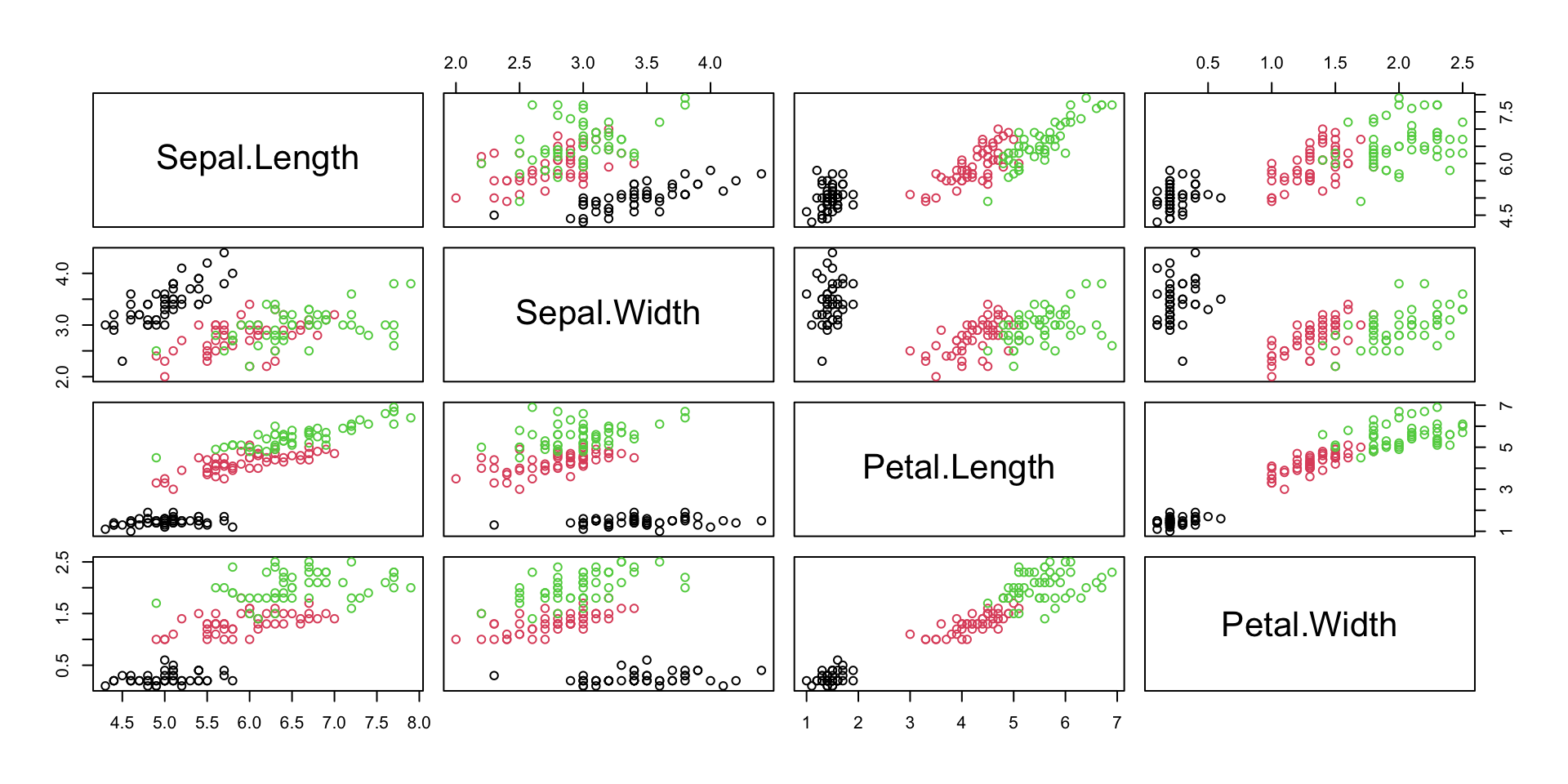
Iris
-
This famous (Fisher’s or Anderson’s) iris data set gives the measurements in centimeters of the following four variables:
- sepal length and width
- petal length and width,
Data set contains the measurements on 50 flowers from each of 3 species of iris.
The species are Iris
setosa,versicolor, andvirginica.
Iris plot
Demo 1
These are the same solution but with swapped labels
Demo 2
These are not the same solution
Demo 3
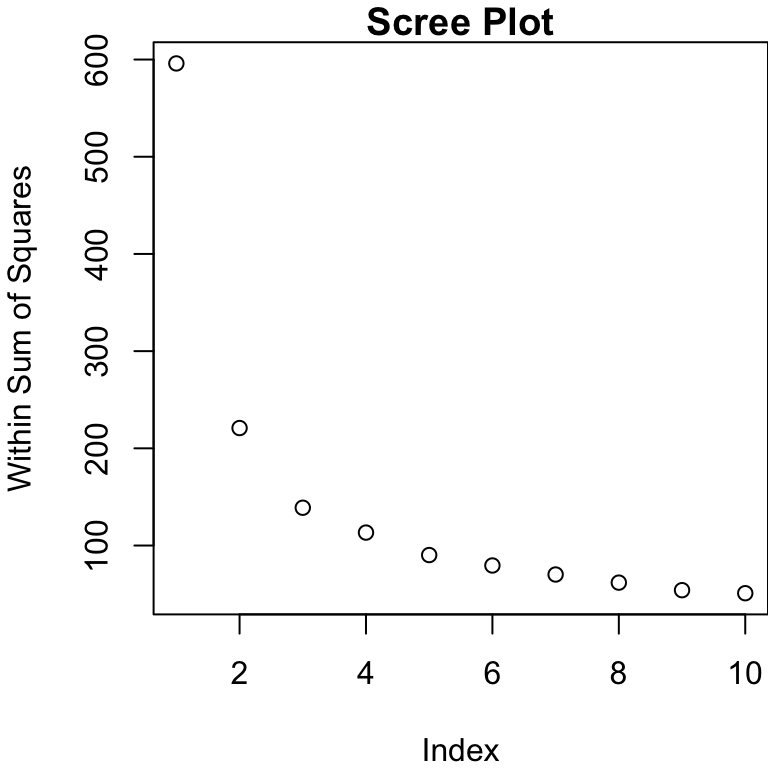
Determining Number of Groups
For hierarchical, we have an argument for the number of groups present in the data (largest jump in dendrogram).
For \(k\)-means, we need to specify \(k\) but can still provide some guidance
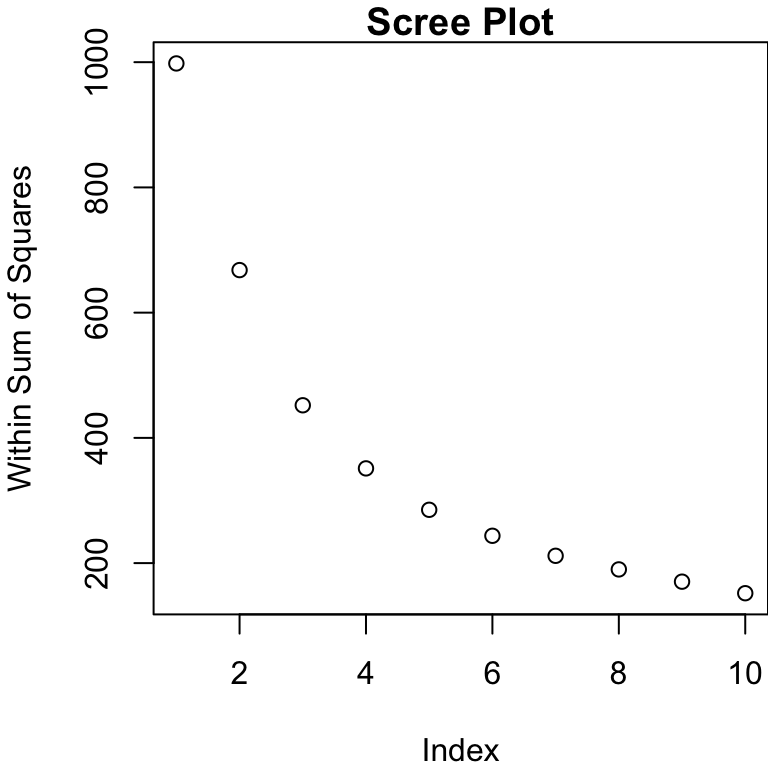
We can plot the WSS (within sum of squares) for differing numbers of groups
WSS
We look for the “elbow” in the so-called “scree” plot.
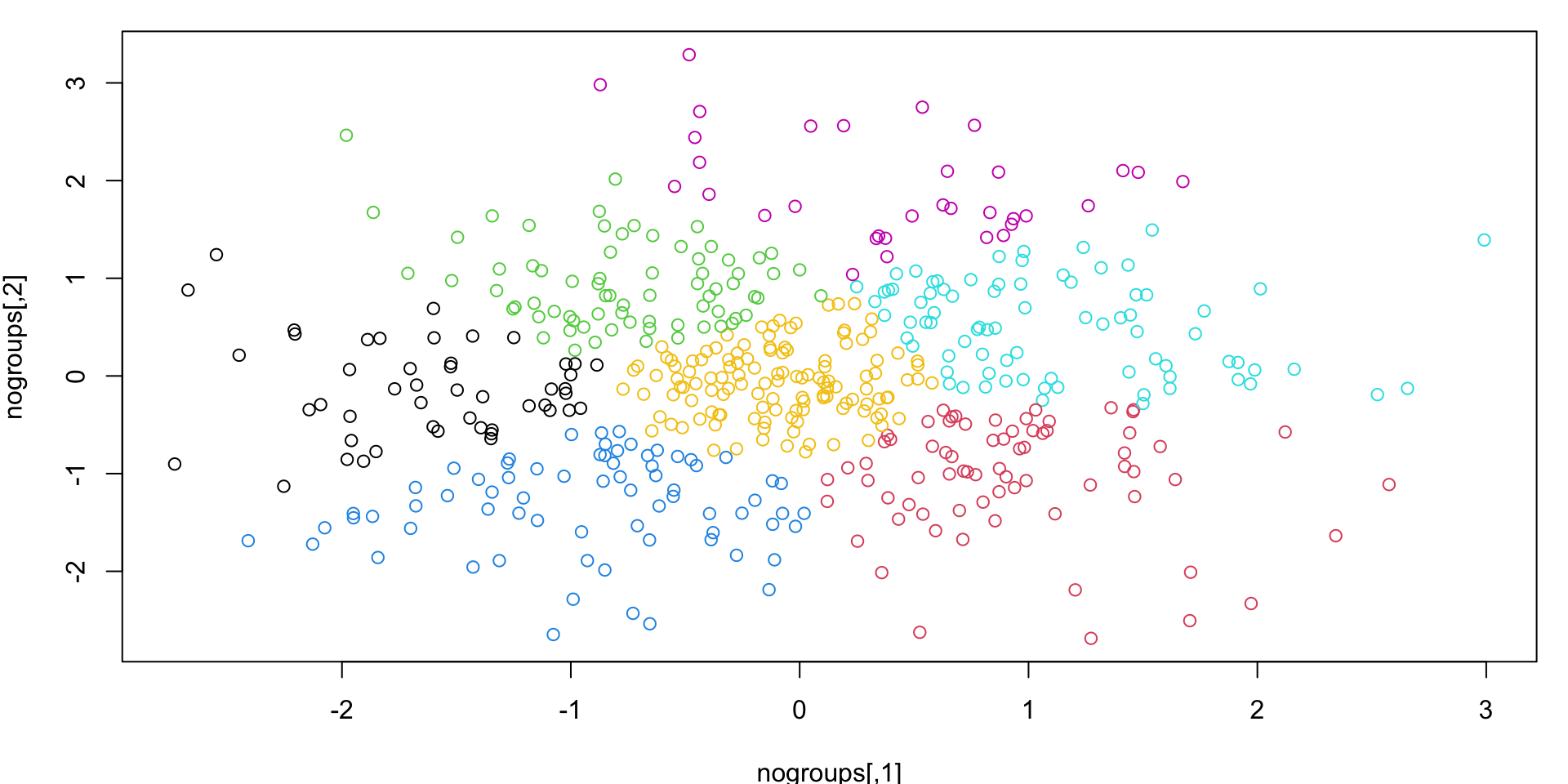
No groups?
Also as previously noted, \(k\)-means can seem like it finds groups, even when they may not exist.

k-means “solution”
K-means with \(k\) = 7 will always return a “solution”



Comments before we begin
Clustering will have an element of subjectivity (some may argue that there are 3 groups in the Old Faithful data set)
It can therefore be hard to assess the results obtained from unsupervised learning methods
For these reasons, clustering algorithms are often tried and tested on data sets for which classes do exist (eg. wine)